What is APOBEC3G?
|
|
|
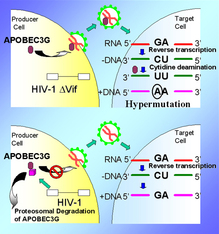
In a small number of people infected with HIV, the virus is naturally suppressed without medical treatment. These people may carry high quantities of a protein called APOBEC3G that disrupts viral replication in cells. APOBEC3G, or "A3" for short, is a protein that sabotages reverse transcription, the process HIV relies on for its replication. This process involves the virus transcribing its singe-stranded RNA genome into double-stranded DNA that is incorporated into the cell's genome. A3 usually stops dormant viruses in the human genome, called endogenous retroviruses, from reawakening and causing infections.
This gene is a member of the cytidine deaminase gene family. It is one of seven related genes or pseudogenes found in a cluster, thought to result from gene duplication, on chromosome 22. Members of the cluster encode proteins that are structurally and functionally related to the C to U RNA-editing cytidine deaminase APOBEC1. It is thought that the proteins may be RNA editing enzymes and have roles in growth or cell cycle control. The protein encoded by this gene has been found to be a specific inhibitor of human immunodeficiency virus-1 (HIV-1) infectivity.
This gene is a member of the cytidine deaminase gene family. It is one of seven related genes or pseudogenes found in a cluster, thought to result from gene duplication, on chromosome 22. Members of the cluster encode proteins that are structurally and functionally related to the C to U RNA-editing cytidine deaminase APOBEC1. It is thought that the proteins may be RNA editing enzymes and have roles in growth or cell cycle control. The protein encoded by this gene has been found to be a specific inhibitor of human immunodeficiency virus-1 (HIV-1) infectivity.
APOBEC3G and HIV
Michael Malim, PhD, of King's College, London, United Kingdom, delivered a state-of-the-art overview of data on the HIV-encoded Vif protein and the cellular enzyme, APOBEC, summarizing our knowledge of the unique interactions between the 2.[1] According to our current understanding, APOBEC is capable of mutagenizing HIV, and Vif destroys APOBEC. The lecture was given in memory of Dr. Bernard Fields, a leading virologist, who pioneered many key concepts in virology and antiviral drug development.
Dr. Malim began by pointing out that controversy in regard to the role of Vif in the HIV life cycle had existed over many years. Indeed, many scientists had argued that Vif was not really essential for virus replication because viruses deleted of the vif gene were still able to replicate efficiently in some cell systems. The reason for this is the presence of a cellular enzyme termed APOBEC3G (APOBEC) that can limit the replication of HIV-1.
When it is present in adequate amounts, the APOBEC enzyme is incorporated into virus particles and attacks newly made HIV RNA or DNA: cytosine (C) residues are changed to uracil (U) residues in viral RNA, or guanosine (G) to adenosine (A) residues in viral DNA, thus yielding a nonactive form of the HIV genome. In contract, Vif, if present in virus particles, will cause the degradation of APOBEC3G, thereby precluding modification of the viral genome and permitting viral replication to proceed. The mechanism through which this is accomplished is the recruitment of cellular ubiquitin ligases and the resultant elimination of APOBEC in proteasomes.
In brief, the HIV Vif protein will prevent APOBEC3G from being encapsulated by virus particles. Although APOBEC3G is a member of a family of cellular enzymes termed cytidine deaminases, it seems to be unique in regard to its ability to interact with and be antagonized by Vif, which is a virus-encoded cytoplasmic protein.
Several workers have shown that many primate cells are resistant to infection by HIV-1. This resistance is mediated by Trim5-alpha, a cellular protein that can interfere with HIV reverse transcription. One mechanism of such resistance may be through the enhanced production of APOBEC3G.
An intriguing question is how simpler retroviruses may have evolved without needing to possess a Vif-like protein. The answer seems to be, in part, that both Vif and APOBEC3G may have coevolved as a means of facilitating -- but not overly so -- the ability of lentiviruses to mediate infection. Accordingly, many scientists believe that Vif and APOBEC normally coexist in equilibrium, because interference with Vif function might increase HIV mutation rates in a way that might actually be deleterious for the virus.
Finally, research has shown that interactions between APOBEC3G and Vif are species-specific. For example, the Vif protein of HIV-1 may be able to neutralize APOBEC3G that is present within human cells but may not be able to efficiently neutralize an equivalent cellular protein that is found in macaque lymphocytes. It is interesting to note that the Vif protein of simian immunodeficiency virus (SIV) can neutralize APOBEC3G of both macaque and human cells, but this virus is also able to infect both cell types. These data may help us to understand the evolutionary process involving Vif and APOBEC.
Hope for Novel Therapeutic Strategies?Interfering with the ability of Vif and APOBEC3G to interact constitutes a rationale for antiviral drug development, and research on such approaches is ongoing in a number of laboratories. Further understanding of this subject is afforded by the fact that viral genomic RNA is normally bound to zinc finger motifs that are present in the viral nucleocapsid protein that plays a large role in viral assembly. APOBEC3G can bind directly to viral RNA, but its ability to do so is dependent on the long intracellular half-life of the latter, and elimination of the zinc finger motifs will compromise this binding activity.
APOBEC3G may be distinguished from other members of the cytidine deaminase family by its zinc fingers, which may be essential for the Vif interaction to take place. These zinc fingers may also represent a potential target for drug development. However, such approaches may prove to be toxic because many cellular proteins also possess zinc fingers, and may also be disrupted by antiviral compounds that target such structures.
Another strategy might be to try to augment levels of APOBEC3G that are normally present inside cells. Although such an approach can be contemplated through genetic approaches, this may not easily translate into efficient antiviral drug development.
An improved analysis and functional dissection of the regulatory processes that govern the Vif-APOBEC3G interaction will hopefully lead to novel ways of dealing with and/or overcoming HIV infection.
Reference
1. Malim MH. Natural resistance to HIV infection: the Vif-APOBEC interaction. Program and abstracts of the 12th Conference on Retroviruses and Opportunistic Infections; February 22-25, 2005; Boston, Massachusetts. Abstract 5.
Dr. Malim began by pointing out that controversy in regard to the role of Vif in the HIV life cycle had existed over many years. Indeed, many scientists had argued that Vif was not really essential for virus replication because viruses deleted of the vif gene were still able to replicate efficiently in some cell systems. The reason for this is the presence of a cellular enzyme termed APOBEC3G (APOBEC) that can limit the replication of HIV-1.
When it is present in adequate amounts, the APOBEC enzyme is incorporated into virus particles and attacks newly made HIV RNA or DNA: cytosine (C) residues are changed to uracil (U) residues in viral RNA, or guanosine (G) to adenosine (A) residues in viral DNA, thus yielding a nonactive form of the HIV genome. In contract, Vif, if present in virus particles, will cause the degradation of APOBEC3G, thereby precluding modification of the viral genome and permitting viral replication to proceed. The mechanism through which this is accomplished is the recruitment of cellular ubiquitin ligases and the resultant elimination of APOBEC in proteasomes.
In brief, the HIV Vif protein will prevent APOBEC3G from being encapsulated by virus particles. Although APOBEC3G is a member of a family of cellular enzymes termed cytidine deaminases, it seems to be unique in regard to its ability to interact with and be antagonized by Vif, which is a virus-encoded cytoplasmic protein.
Several workers have shown that many primate cells are resistant to infection by HIV-1. This resistance is mediated by Trim5-alpha, a cellular protein that can interfere with HIV reverse transcription. One mechanism of such resistance may be through the enhanced production of APOBEC3G.
An intriguing question is how simpler retroviruses may have evolved without needing to possess a Vif-like protein. The answer seems to be, in part, that both Vif and APOBEC3G may have coevolved as a means of facilitating -- but not overly so -- the ability of lentiviruses to mediate infection. Accordingly, many scientists believe that Vif and APOBEC normally coexist in equilibrium, because interference with Vif function might increase HIV mutation rates in a way that might actually be deleterious for the virus.
Finally, research has shown that interactions between APOBEC3G and Vif are species-specific. For example, the Vif protein of HIV-1 may be able to neutralize APOBEC3G that is present within human cells but may not be able to efficiently neutralize an equivalent cellular protein that is found in macaque lymphocytes. It is interesting to note that the Vif protein of simian immunodeficiency virus (SIV) can neutralize APOBEC3G of both macaque and human cells, but this virus is also able to infect both cell types. These data may help us to understand the evolutionary process involving Vif and APOBEC.
Hope for Novel Therapeutic Strategies?Interfering with the ability of Vif and APOBEC3G to interact constitutes a rationale for antiviral drug development, and research on such approaches is ongoing in a number of laboratories. Further understanding of this subject is afforded by the fact that viral genomic RNA is normally bound to zinc finger motifs that are present in the viral nucleocapsid protein that plays a large role in viral assembly. APOBEC3G can bind directly to viral RNA, but its ability to do so is dependent on the long intracellular half-life of the latter, and elimination of the zinc finger motifs will compromise this binding activity.
APOBEC3G may be distinguished from other members of the cytidine deaminase family by its zinc fingers, which may be essential for the Vif interaction to take place. These zinc fingers may also represent a potential target for drug development. However, such approaches may prove to be toxic because many cellular proteins also possess zinc fingers, and may also be disrupted by antiviral compounds that target such structures.
Another strategy might be to try to augment levels of APOBEC3G that are normally present inside cells. Although such an approach can be contemplated through genetic approaches, this may not easily translate into efficient antiviral drug development.
An improved analysis and functional dissection of the regulatory processes that govern the Vif-APOBEC3G interaction will hopefully lead to novel ways of dealing with and/or overcoming HIV infection.
Reference
1. Malim MH. Natural resistance to HIV infection: the Vif-APOBEC interaction. Program and abstracts of the 12th Conference on Retroviruses and Opportunistic Infections; February 22-25, 2005; Boston, Massachusetts. Abstract 5.
Summary for APOBEC3G Gene:
This gene is a member of the cytidine deaminase gene family. It is one of seven related genes or pseudogenes found in a cluster, thought to result from gene duplication, on chromosome 22. Members of the cluster encode proteins that are structurally and functionally related to the C to U RNA-editing cytidine deaminase APOBEC1. It is thought that the proteins may be RNA editing enzymes and have roles in growth or cell cycle control. The protein encoded by this gene has been found to be a specific inhibitor of human immunodeficiency virus-1 (HIV-1) infectivity. (provided by RefSeq, Jul 2008)
Summary for APOBEC3G Gene:
APOBEC3G (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G) is a protein-coding gene. Diseases associated with APOBEC3G include human immunodeficiency virus infectious disease, and hiv-1. GO annotations related to this gene include cytidine deaminase activity and RNA binding.
DNA deaminase (cytidine deaminase) which acts as an inhibitor of retrovirus replication and retrotransposon mobility via deaminase-dependent and -independent mechanisms. Exhibits potent antiviral activity against vif-deficient HIV-1. After the penetration of retroviral nucleocapsids into target cells of infection and the initiation of reverse transcription, it can induce the conversion of cytosine to uracil in the minus-sense single-strand viral DNA, leading to G-to-A hypermutations in the subsequent plus-strand viral DNA. The resultant detrimental levels of mutations in the proviral genome, along with a deamination-independent mechanism that works prior to the proviral integration, together exert efficient antiretroviral effects in infected target cells. Selectively targets single-stranded DNA and does not deaminate double-stranded DNA or single-or double-stranded RNA. Exhibits antiviral activity also against simian immunodeficiency viruses (SIVs), hepatitis B virus (HBV), equine infectious anemia virus (EIAV), xenotropic MuLV-related virus (XMRV) and simian foamy virus (SFV). May inhibit the mobility of LTR and non-LTR retrotransposons
This gene is a member of the cytidine deaminase gene family. It is one of seven related genes or pseudogenes found in a cluster, thought to result from gene duplication, on chromosome 22. Members of the cluster encode proteins that are structurally and functionally related to the C to U RNA-editing cytidine deaminase APOBEC1. It is thought that the proteins may be RNA editing enzymes and have roles in growth or cell cycle control. The protein encoded by this gene has been found to be a specific inhibitor of human immunodeficiency virus-1 (HIV-1) infectivity. (provided by RefSeq, Jul 2008)
Summary for APOBEC3G Gene:
APOBEC3G (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G) is a protein-coding gene. Diseases associated with APOBEC3G include human immunodeficiency virus infectious disease, and hiv-1. GO annotations related to this gene include cytidine deaminase activity and RNA binding.
DNA deaminase (cytidine deaminase) which acts as an inhibitor of retrovirus replication and retrotransposon mobility via deaminase-dependent and -independent mechanisms. Exhibits potent antiviral activity against vif-deficient HIV-1. After the penetration of retroviral nucleocapsids into target cells of infection and the initiation of reverse transcription, it can induce the conversion of cytosine to uracil in the minus-sense single-strand viral DNA, leading to G-to-A hypermutations in the subsequent plus-strand viral DNA. The resultant detrimental levels of mutations in the proviral genome, along with a deamination-independent mechanism that works prior to the proviral integration, together exert efficient antiretroviral effects in infected target cells. Selectively targets single-stranded DNA and does not deaminate double-stranded DNA or single-or double-stranded RNA. Exhibits antiviral activity also against simian immunodeficiency viruses (SIVs), hepatitis B virus (HBV), equine infectious anemia virus (EIAV), xenotropic MuLV-related virus (XMRV) and simian foamy virus (SFV). May inhibit the mobility of LTR and non-LTR retrotransposons

