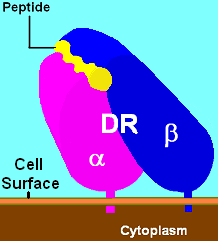
What is DRB1*13 and DQB1*6?
|
The HIV "Elites:" Viral Controllers and Their Genes
By Jeffrey Laurence, M.D.
 amfAR grantee Dr. Steven Deeks
amfAR grantee Dr. Steven Deeks
December 13, 2010 - For more than a decade, Dr. Steven Deeks of the University of California, San Francisco, has been trying to solve an important puzzle. How are a small number of HIV-infected individuals who are not taking antiretroviral therapy (ART) able to maintain low viral loads and near normal CD4 T-cell counts? The answer to this critical question may provide a clue in the design of an effective AIDS vaccine, a goal that has eluded researchers for decades. With support from amfAR, Dr. Deeks and his colleagues at UCSF have assembled a large cohort of these individuals, known as “elite controllers,” and is collaborating with other groups to determine how the immune system keeps their virus in check.
In a recent study led by Drs. April Ferre and Barbara Shacklett at the University of California, Davis, Deeks and his colleagues obtained blood samples and biopsies of rectal tissue from 52 HIV-positive patients: 28 elite controllers, 14 “noncontrollers” who were not on treatment, and 10 noncontrollers on ART who maintained undetectable viral loads. The biopsies were necessary as the intestinal tract is a key site for HIV transmission, growth, and depletion of T cells, and has often been overlooked in studies of elite controllers.
The researchers found that many study participants, both controllers and noncontrollers, had similar levels of HIV-specific CD4 T cells—immune cells that direct the response of the immune system to the virus. But the controllers had “unusually strong” CD4 T cells in their rectal tissue, cells capable of recognizing proteins known as Gag proteins, which form the core of the virus. In some controllers, more than one CD4 T cell in every 10 counted proved to be HIV-specific. This level was much greater than what was found in participants who required ART to keep circulating levels of virus low. Controllers also had higher levels of multi-functional CD4 cells in rectal tissue than noncontrollers or those on ART. Along with these super-functional CD4 T cells, another type of immune cell known as the CD8 T cell was also present at much higher levels in controllers. This finding makes sense, since CD8 T cells are under the control of the CD4 T cells that are especially virulent in controllers.
In exploring possible genetic explanations for viral control, the researchers found that controllers with the strongest HIV-specific CD4 cells had at least one of two genes associated with defining tissue type: DRB1*13 and DQB1*6. These genes have previously been linked to HIV-positive individuals called nonprogressors, who do not progress to AIDS despite lack of treatment. Deeks and his colleagues have broadened our understanding of this phenomenon with their finding that those lucky enough to possess both DRB1*13 and DQB1*6 have T cells with super-strength in recognizing HIV.
The team concluded that the preservation and expansion of both HIV-specific CD4 and CD8 T cells may be key mechanisms by which HIV controllers keep their virus in check. Their findings suggest that HIV vaccines and therapies aimed at enhancing these intestines-associated T-cell responses should be actively pursued.
Dr. Laurence is amfAR’s senior scientific consultant.
Article: http://www.amfar.org/articles/in-the-lab/2010/the-hiv-%E2%80%9Celites-%E2%80%9D-viral-controllers-and-their-genes-(december-2010)/
In a recent study led by Drs. April Ferre and Barbara Shacklett at the University of California, Davis, Deeks and his colleagues obtained blood samples and biopsies of rectal tissue from 52 HIV-positive patients: 28 elite controllers, 14 “noncontrollers” who were not on treatment, and 10 noncontrollers on ART who maintained undetectable viral loads. The biopsies were necessary as the intestinal tract is a key site for HIV transmission, growth, and depletion of T cells, and has often been overlooked in studies of elite controllers.
The researchers found that many study participants, both controllers and noncontrollers, had similar levels of HIV-specific CD4 T cells—immune cells that direct the response of the immune system to the virus. But the controllers had “unusually strong” CD4 T cells in their rectal tissue, cells capable of recognizing proteins known as Gag proteins, which form the core of the virus. In some controllers, more than one CD4 T cell in every 10 counted proved to be HIV-specific. This level was much greater than what was found in participants who required ART to keep circulating levels of virus low. Controllers also had higher levels of multi-functional CD4 cells in rectal tissue than noncontrollers or those on ART. Along with these super-functional CD4 T cells, another type of immune cell known as the CD8 T cell was also present at much higher levels in controllers. This finding makes sense, since CD8 T cells are under the control of the CD4 T cells that are especially virulent in controllers.
In exploring possible genetic explanations for viral control, the researchers found that controllers with the strongest HIV-specific CD4 cells had at least one of two genes associated with defining tissue type: DRB1*13 and DQB1*6. These genes have previously been linked to HIV-positive individuals called nonprogressors, who do not progress to AIDS despite lack of treatment. Deeks and his colleagues have broadened our understanding of this phenomenon with their finding that those lucky enough to possess both DRB1*13 and DQB1*6 have T cells with super-strength in recognizing HIV.
The team concluded that the preservation and expansion of both HIV-specific CD4 and CD8 T cells may be key mechanisms by which HIV controllers keep their virus in check. Their findings suggest that HIV vaccines and therapies aimed at enhancing these intestines-associated T-cell responses should be actively pursued.
Dr. Laurence is amfAR’s senior scientific consultant.
Article: http://www.amfar.org/articles/in-the-lab/2010/the-hiv-%E2%80%9Celites-%E2%80%9D-viral-controllers-and-their-genes-(december-2010)/
HIV Controllers with HLA-DRB1*13 and HLA-DQB1*06 Alleles Have Strong, Polyfunctional Mucosal CD4+ T-Cell Responses▿†
ABSTRACTA small percentage of human immunodeficiency virus (HIV)-infected individuals, termed elite controllers, are able to spontaneously control HIV replication in blood. As the gastrointestinal mucosa is an important site of HIV transmission and replication as well as CD4+ T-cell depletion, it is important to understand the nature of the immune responses occurring in this compartment. Although the role of the HIV-specific CD8+ T-cell responses in mucosal tissues has been described, few studies have investigated the role of mucosal HIV-specific CD4+ T cells. In this study, we assessed HIV-specific CD4+ T-cell responses in the rectal mucosa of 28 “controllers” (viral load [VL] of <2,000 copies/ml), 14 “noncontrollers” (VL of >10,000 copies/ml), and 10 individuals on highly active antiretroviral therapy (HAART) (VL of <75 copies/ml). Controllers had higher-magnitude Gag-specific mucosal CD4+ T-cell responses than individuals on HAART (P < 0.05), as measured by their ability to produce gamma interferon (IFN-γ), interleukin-2 (IL-2), tumor necrosis factor alpha (TNF-α), and macrophage inflammatory protein 1β (MIP-1β). The frequency of polyfunctional mucosal CD4+ T cells was also higher in controllers than in noncontrollers or individuals on HAART (P < 0.05). Controllers with the strongest HIV-specific CD4+ T-cell responses possessed class II HLA alleles, HLA-DRB1*13 and/or HLA-DQB1*06, previously associated with a nonprogression phenotype. Strikingly, individuals with both HLA-DRB1*13 and HLA-DQB1*06 had highly polyfunctional mucosal CD4+ T cells compared to individuals with HLA-DQB1*06 alone or other class II alleles. The frequency of polyfunctional CD4+ T cells in rectal mucosa positively correlated with the magnitude of the mucosal CD8+ T-cell response (Spearman's r = 0.43, P = 0.005), suggesting that increased CD4+ T-cell “help” may be important in maintaining strong CD8+ T-cell responses in the gut of HIV controllers.
The study of individuals who are able to achieve durable control over human immunodeficiency virus (HIV) replication in the absence of antiretroviral therapy has become increasingly important in light of recent vaccine trials which have failed to induce protective immunity. Understanding the correlates of protection in these HIV “controllers” would aid in both the rational design of potential vaccine candidates and the testing of their efficacy.
Cell-mediated immune responses, in particular HIV-specific CD8+ T-cell responses, have been shown to be critical in decreasing the initial viremia after acute infection and determining the chronic infection set point (32, 33, 43). These responses have proven to be “too little and too late” to prevent the establishment of chronic infection; however, studies of HIV-specific CD8+ T-cell responses in controllers suggest that strong, polyfunctional responses may be important in long-term virologic control (1, 6, 11, 15, 16, 44). In addition, the association of controller status with class I HLA alleles, predominantly HLA-B27 and HLA-B57, has been well documented in multiple cohorts (2, 9, 13, 39).
The maintenance of robust HIV-specific CD4+ T-cell responses may also favor long-term control of HIV replication in untreated persons. One of the primary roles of CD4+ T cells is to provide “help” to CD8+ T cells. Many studies have shown that proper functioning of CD4+ T cells is necessary for high-quality antigen-specific CD8+ T-cell responses (3, 28, 29, 38, 58). HIV-specific CD4+ T cells in the peripheral blood of long-term nonprogressors (LTNP) have been shown to be polyfunctional, producing both gamma interferon (IFN-γ) and interleukin-2 (IL-2), whereas those from progressors tend to be monofunctional, secreting only IFN-γ (7, 14, 21, 22, 44). Additionally, the ability of CD4+ T cells to proliferate appears to be preserved in controllers (46, 56). The maturation status of CD4+ T cells that function in the context of HIV infection may also be important, as individuals who are able to preserve central memory T cells and sustain an activated effector memory CD4+ T-cell population are better able to suppress viral replication (45). Much less is known about the association of class II HLA alleles and controller status; however, a few studies have cited a relationship between HLA-DRB1*13 and/or HLA-DQB1*06 and HIV control (10, 31, 36, 55).
Previously, we have shown that CD8+ T cells from the rectal mucosa of controllers with protective class I alleles (HLA-B13, -B27, -B57, -B58, and -B81) are highly polyfunctional compared to CD8+ T cells from either controllers or noncontrollers (NC) lacking these alleles (16). Additionally, we found that mucosal CD8+ T-cell responses from individuals who had protective class I alleles in combination with the class II alleles HLA-DRB1*13 and/or HLA-DQB1*06 were of greater magnitude than mucosal CD8+ T-cell responses from those who had protective class I alleles alone (16). Therefore, we wanted to specifically examine HIV-specific mucosal CD4+ T-cell responses among controllers with and without these potentially protective class II HLA alleles.
Our hypothesis was that controllers, particularly those who possessed HLA-DRB1*13 and/or HLA-DQB1*06, would have more robust and polyfunctional HIV-specific CD4+ T-cell responses than noncontrollers or subjects on highly active antiretroviral therapy (HAART) and that these responses would correlate with strong CD8+ T-cell responses in the same individuals. We found that, indeed, controllers generally had stronger HIV-specific CD4+ T-cell responses than other groups in rectal mucosa and that, among controllers, those with the haplotype HLA-DRB1*13/HLA-DQB1*06 had particularly high percentages of polyfunctional HIV-specific CD4+ T cells in rectal mucosa. Furthermore, the proportion of polyfunctional mucosal CD4+ T cells directly correlated with the total magnitude of the mucosal CD8+ T-cell response. These data collectively suggest that the preservation or expansion of both HIV-specific CD4+ and CD8+ T cells may be an important mechanism whereby certain controllers maintain durable control of HIV replication.
Previous SectionNext SectionMATERIALS AND METHODSSubjects and tissue collection.Subjects were recruited through ongoing studies of chronic HIV infection at San Francisco General Hospital and the Center for AIDS Research, Education, and Services (CARES) Clinic in Sacramento, CA, and have been described previously (16). Subjects were classified in one of five categories based on viral load (VL) measurements and treatment status, as described by Deeks and Walker (12): elite controllers (EC; VL of <75 copies/ml, off therapy, n = 17), viremic controllers (VC; VL of <2,000 copies/ml, off therapy, n = 11), noncontrollers (NC; VL of >10,000 copies, off therapy, n = 14), highly active antiretroviral therapy (HAART)-suppressed subjects (VL of <75 copies, on HAART, n = 10), and HIV-negative controls (n = 8). Written informed consent for phlebotomy and flexible sigmoidoscopy was obtained from all subjects in accordance with the Declaration of Helsinki, and study protocols were approved by the Institutional Review Board, University of California—Davis, and the Committee on Human Subjects Research, University of California—San Francisco.
Approximately 20 ml of blood was obtained by sterile venipuncture and collected into tubes containing EDTA. Rectal biopsy tissue (20 to 25 pieces) was obtained by flexible sigmoidoscopy at 10 cm from the anal verge using a sigmoidoscope equipped with a biopsy channel and single-use biopsy forceps. This procedure has been well documented to cause only minimal discomfort and provide enough cells to perform cellular immunology assays (mean of 10 × 106 cells) (11, 16, 49, 51). Tissue was placed in RPMI 1640 supplemented with 15% fetal calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and l-glutamine (2 nM), herein referred to as R15. Specimens were immediately transported to the University of California—Davis for same-day processing.
Peripheral blood and rectal biopsy tissue processing.Peripheral blood was layered onto a Ficoll-Hypaque (Pfizer, New York, NY) density gradient to isolate mononuclear cells (PBMC). Rectal biopsy tissue was processed according to a previously published protocol designed to maximize viable lymphocyte yield (11, 16, 49, 51). Briefly, biopsy tissue was subjected to three rounds of collagenase type II digestion (0.5 mg/ml; Sigma-Aldrich, St. Louis, MO), followed by mechanical disruption using an 18-gauge blunt-end needle and passage through a 70-μm cell strainer. Cells were then washed in R15 and layered on a 35%/65% Percoll gradient (Sigma-Aldrich). Mononuclear cells were harvested from both the medium-35% Percoll interface and the 35%/65% Percoll interface to maximize yield. PBMC and rectal mononuclear cells (RMC) were rested overnight at 37°C and 5% CO2, and piperacillin-tazobactam (Zosyn, 0.5 mg/ml; Wyeth-Ayerst, Princeton, NJ) was added to RMC cultures to prevent bacterial growth.
HLA class II typing.Genomic DNA was isolated from 5 × 106 to 8 × 106 PBMC by using a QIAamp DNA blood minikit (Qiagen, Valencia, CA) and quantified by spectrophotometer. Low-resolution HLA-DR and HLA-DQ typing was performed using PCR and a DR/DQ 2T locus SSP (sequence-specific primer) Unitray kit (Invitrogen, Carlsbad, CA). PCR products were resolved by electrophoresis on a 2% agarose gel, photographed, and analyzed using UniMatch Plus software (Invitrogen).
Antibodies and peptide pools.Fluorochrome-labeled monoclonal antibodies to CD3 (clone UCHT-1), CD107a (clone H4A3), gamma interferon (IFN-γ; clone B27), macrophage inflammatory protein 1β (MIP-1β; clone D21-1351), tumor necrosis factor alpha (TNF-α; clone MAb11), and interleukin-2 (IL-2; clone 5344.111) and unlabeled antibodies to CD28 (clone L293) and CD49d (clone L25) were purchased from BD Biosciences (San Jose, CA). Fluorochrome-labeled antibodies to CD4 (clone SFCI12T4D11) and CD8 (clone SK1) were purchased from Beckman Coulter (Fullerton, CA) and Invitrogen, respectively. All antibodies were titrated to determine optimum concentrations for assay conditions (data not shown). HIV Gag (p55, HXB2 sequence) peptide pools containing 15-mer peptides overlapping by 11 amino acids (aa) were purchased from BD Biosciences.
Intracellular cytokine staining and flow cytometry.Intracellular cytokine staining of newly isolated PBMC or RMC without prior expansion was performed as previously described (16). Briefly, 1 × 106 to 2 × 106 PBMC or RMC were stimulated for 5 h with the HIV Gag p55 pool (3.5 μg/ml) in the presence of anti-CD28 (2.5 μg/ml) and anti-CD49d (5 μg/ml) costimulatory antibodies, anti-CD107a, Golgi Stop (1 μM; BD Biosciences), and brefeldin A (5 μg/ml; Sigma-Aldrich). Dimethyl sulfoxide (DMSO) and Staphylococcus enterotoxin B (SEB; 5 μg/ml) served as negative and positive controls, respectively. Cells were then stained with antibodies to surface antigens CD4 and CD8 and with 7-amino-actinomycin D (7-AAD; BD Biosciences) to label dead cells. Thereafter, all buffers and reagents contained actinomycin D to saturate sites with the potential to bind 7-AAD. Cells were fixed in 4% paraformaldehyde and then permeabilized using FACS Perm 2 (BD Biosciences). Intracellular cytokine staining for CD3, IFN-γ, IL-2, MIP-1β, and TNF-α followed permeabilization. Finally, the cells were placed in 1% paraformaldehyde for at least 1 h, but no longer than 24 h, before being read on an LSRII flow cytometer equipped with a 405-, 488-, and 643-nm laser (BD Biosciences).
Polyclonal CD4+ T-cell expansion of peripheral blood and rectal mononuclear cells.Polyclonal expansion was performed in order to obtain sufficient CD4+ T cells for epitope mapping using the enzyme-linked immunospot (ELISpot) method (47). This approach has been shown to expand T cells nonspecifically without significantly altering T-cell receptor (TCR) clonotypes, patterns of epitope recognition, or the ability of T cells to respond to peptide stimulation by secreting IFN-γ (4, 25, 26). It should be noted that, in the present study, polyclonal expansion was utilized only to generate sufficient cells to map the peptide specificity of responses that had previously been identified by intracellular cytokine staining of fresh PBMC and RMC. One million PBMC or RMC were placed in 2 ml R15 in a 24-well plate with 1 μg/ml CD3/CD8-bispecific antibody (generously provided by Johnson Wong, Harvard University), which preferentially expands CD4+ T cells while promoting apoptosis in CD8+ T cells (57). PBMC and RMC cultures were supplemented with recombinant IL-2 (rIL-2) (50 U/ml; R&D Systems, Minneapolis, MN).
When initial attempts to culture RMC (but not PBMC) gave a low success rate, we supplemented cultures with 1 ng/ml human recombinant IL-7 (R&D Systems, Minneapolis, MN), based on a previously published protocol. This cytokine promotes survival of memory T cells, in part by inactivating proapoptotic pathways (34). Amphotericin B (1.25 μg/ml; MP Biomedicals, Solon, OH) and piperacillin-tazobactam (Zosyn, 0.5 μg/ml; Wyeth-Ayerst, Princeton, NJ) were added to prevent fungal and bacterial growth, respectively. After the first week, IL-2 (and IL-7 for RMC cultures) was added twice weekly, as were additional amphotericin B and piperacillin-tazobactam, to compensate for any added R15 volume during the expansion. Two to 3 days following the initial CD3/CD8 antibody stimulation, 2 × 106 irradiated heterologous PBMC (5,000 rad) from a seronegative donor were added to cultures. Cultures were expanded and maintained for 4 to 8 weeks. After 3 weeks of culture, cells were restimulated with 0.1 μg/ml 12F6 anti-CD3 antibody (also provided by Johnson Wong, Harvard University).
CD4+ T-cell response mapping.By 4 to 5 weeks, polyclonally expanded cultures were composed of primarily CD4+ T cells; however, approximately half of the cultures included 20 to 40% CD8+ T cells. Therefore, CD8+ T cells were depleted from cultures by using magnetic bead separation prior to epitope mapping. Anti-CD8 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) were used to label CD8+ T cells, which were then removed from the CD4+ T cells by using a MACS system (Miltenyi Biotec) with an LD column.
CD4+ T-cell response mapping was performed using an IFN-γ ELISpot assay as previously described for CD8+ T-cell response mapping (50). Briefly, pools of HIV HXB2 consensus clade B Gag, Env, and Nef peptides (15-mer peptides overlapping by 11; NIH AIDS Research and Reference Reagent Program, Rockville, MD) were created so that each peptide was contained in exactly two pools. This allowed the creation of a peptide matrix for screening HIV peptides. Peptides common to pools positive for a CD4+ T-cell response (>50 spot-forming cells [SFC]/million after the subtraction of background) were then tested individually using a standard IFN-γ ELISpot assay.
Data analysis.Flow cytometry data were analyzed with FlowJo software (TreeStar, Ashland, OR). Boolean gate analysis allowed the separation of CD4+ T-cell responses into 16 individual combinations of the four functional parameters (IFN-γ, IL-2, MIP-1β, and TNF-α). Data generated from Boolean gate analysis were then visualized in SPICE software (v.4.2.2; Mario Roederer, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD). Standard bivariate plots were also constructed to confirm the placement of individual Boolean gates.
IFN-γ ELISpots were read on an AID plate reader (Autoimmun Diagnostika GMBH, Straßerg, Germany), and pool- or peptide-specific CD4+ T-cell responses were quantified as SFC/million after the subtraction of background. Responses of >50 SFC/million were considered positive.
Statistical analysis.CD4+ T-cell flow cytometry data were analyzed using a previously described formula which considers whether or not antigen-specific responses differ significantly from background (11, 16). This step was important for evaluating mucosal T-cell responses, as they are highly responsive to costimulation (data not shown) and can produce low levels of cytokines and chemokines in the absence of antigen-specific stimulation. The formula assumes a Poisson distribution and takes into account the actual number of collected events rather than percentages of responding cells, as the lymphocyte yield from rectal mucosa tends to be low. When a determination that antigen-specific values were significantly different from background was made, net responses were then calculated by subtracting unstimulated control values from antigen-specific values. Responses that were not determined to be significantly different from background were dropped (11, 16).
For response data, comparisons between two groups were made using a two-tailed Mann-Whitney test in GraphPad Prism software (GraphPad Software, San Diego, CA). Comparisons between polyfunctionality pie charts utilized a permutation test based on χ2 statistics, and comparisons between two individual functional categories employed a Wilcoxon rank test (SPICE Software). A two-tailed Fisher exact test was used for comparing the distributions of HLA-DRB1*13 and HLA-DQB1*06 among subject groups. Correlations between two variables were done using the Spearman correlation, and linear regression was used to graph a best-fit line to the data (GraphPad Prism Software).
Previous SectionNext SectionRESULTSBaseline characteristics.The individuals in this study have been described in an earlier report (16). Briefly, 52 HIV-positive individuals were assigned to one of four groups, as described in Materials and Methods: elite controllers, viremic controllers, noncontrollers, and HAART-suppressed patients. Eight HIV-negative subjects were included as controls.
The percentage of CD4+ T cells as a proportion of CD3+ T cells in peripheral blood and rectal mucosa was previously determined by flow cytometry (16). Elite and viremic controllers had significantly higher percentages of CD4+ T cells in rectal mucosa than noncontrollers (medians of 42.2% and 30.1% versus 15.1%, respectively; P < 0.05); rectal CD4+ percentages in controllers were comparable to those in subjects on HAART (median of 45.8%). All HIV-positive individuals had lower percentages of CD4+ T cells than HIV-negative controls (median of 73%; P < 0.01) (16).
High-magnitude HIV-specific CD4+ T-cell responses in rectal mucosa.The ability of CD4+ T cells to respond to HIV Gag stimulation by producing IFN-γ, IL-2, MIP-1β, and/or TNF-α was measured by flow cytometry using fresh PBMC and RMC without prior expansion (see reference 16 for gating strategy). CD4+ T-cell responses were generally much higher, in terms of total percentage of responding cells, in rectal mucosa than peripheral blood. This likely reflects the predominance of antigen-experienced memory cells in rectal mucosa and was previously observed for HIV-specific CD8+ T cells (16). The median total HIV-specific mucosal CD4+ T-cell response (cells positive for one or more of the above-described functions, all responses combined) was significantly higher in elite controllers than in individuals on HAART (Fig. 1 A) (3.4% versus 0.4%, respectively; P = 0.045). While some elite controllers had unusually strong Gag-specific mucosal CD4+ T-cell responses (as high as 13%), responses within the group were highly heterogeneous, and therefore the median total CD4+ T-cell responses were not significantly different between elite controllers and noncontrollers (3.4% versus 2.3%, respectively; P = 0.21).
View larger version:FIG. 1. HIV Gag-specific CD4+ T-cell responses in rectal mucosa (A to C) and peripheral blood (D to F). Panels A and D show the total percentages of CD4+ T cells capable of producing IFN-γ, IL-2, MIP-1β, and/or TNF-α in response to HIV Gag stimulation in rectal mucosa (EC versus subjects on HAART, P = 0.045) (A) and peripheral blood (D). Panels B and C show significant differences in CD4+ T-cell cytokine/chemokine expression in rectal mucosa between groups: IFN-γ (EC versus NC, P = 0.031; EC versus subjects on HAART, P = 0.035; VC versus NC, P = 0.0005; VC versus subjects on HAART, P = 0.004) (B); MIP-1β (EC versus NC, P = 0.004; EC versus subjects on HAART, P = 0.016; VC versus NC, P = 0.012) (C). Panels E and F show significant differences in CD4+ T-cell cytokine expression in peripheral blood between groups: IL-2 (EC versus NC, P = 0.008; EC versus subjects on HAART, P = 0.018; VC versus NC, P = 0.003; VC versus subjects on HAART, P = 0.001) (E); TNF-α (VC versus NC, P = 0.019; VC versus subjects on HAART, P = 0.006) (F). Horizontal bars represent the median response for each group. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
While predominantly tissue-specific differences (i.e., between peripheral blood and rectal mucosa) were found in the overall magnitude of HIV-specific CD4+ T-cell responses, differences were found in CD4+ T-cell responses between subject groups when assessed for individual functions. Elite controllers and viremic controllers had higher frequencies of mucosal CD4+ T cells producing IFN-γ (Fig. 1B) and MIP-1β (Fig. 1C) than either noncontrollers (NC) or HAART-suppressed individuals.
In general, <0.5% of peripheral blood CD4+ T cells from controllers, noncontrollers, or HAART-suppressed subjects responded in any way to Gag stimulation (Fig. 1D). There was no consistent difference the in ability of mucosal CD4+ T cells to produce IL-2 or TNF-α across subject groups; however, controllers generally had higher levels of IL-2-producing cells in peripheral blood (Fig. 1E), as previously described by our group and others (7, 14, 21, 44), as well as higher levels of TNF-α (Fig. 1F).
HIV controllers have complex Gag-specific mucosal CD4+ T-cell responses.Boolean gate analysis was used to assess the complexity of the HIV-specific CD4+ T-cell response by looking at all possible combinations of four functional parameters: IFN-γ, IL-2, TNF-α, and MIP-1β. Cells that simultaneously expressed three or four cytokines in response to HIV peptide stimulation were considered “polyfunctional.” As there were no significant differences in polyfunctionality between elite and viremic controllers (data not shown), the two groups were combined into a single “controller” group in order to simplify the analysis. Controllers showed a trend toward a mucosal CD4+ T-cell response that was overall more polyfunctional than that for noncontrollers, as represented by the pie charts in Fig. 2 A (P = 0.051). Broken down into the 16 separate functional categories, controllers had higher frequencies of four-function mucosal CD4+ T cells than noncontrollers and more three-function CD4+ T cells (primarily IFN-γ+ MIP-1β+ TNF-α+ and lacking IL-2) than noncontrollers or individuals on HAART (Fig. 2A) (P < 0.05). Interestingly, controllers showed high levels of MIP-1β single positive cells in rectal mucosa, while noncontrollers had high levels of TNF-α single positive cells, compared to levels in other subject groups (P < 0.05).
View larger version:FIG. 2. HIV Gag-specific CD4+ T-cell polyfunctional responses in rectal mucosa and peripheral blood across subject groups. CD4+ T-cell polyfunctional responses in the rectal mucosa (A) and peripheral blood (B) of controllers, noncontrollers, and HAART-suppressed individuals. Colors in the pie charts represent 4-function cells (black), 3-function cells (purple), 2-function cells (dark blue), and 1-function cells (light blue). Values in the pie charts are the median total CD4+ T-cell response within each subject group. The bar charts below show the responses in controllers, noncontrollers, and HAART-suppressed individuals, broken down into individual functional categories using interquartile ranges. Asterisks below the bar charts show significant differences (P < 0.05; Wilcoxon rank test) between controllers and noncontrollers (red) or subjects on HAART (green).
The response complexity in peripheral blood of HIV controllers appeared to be more polyfunctional than that in noncontrollers or HAART-suppressed subjects, but these differences did not reach statistical significance (Fig. 2B). Similar to responses seen in rectal mucosa, controllers had higher percentages of Gag-specific CD4+ T cells capable of three or four functions than noncontrollers (P < 0.05 for peripheral blood); however, the dominant three-function category (in terms of median response) in blood was IFN-γ+ IL-2+ TNF-α+, rather than IFN-γ+ MIP-1β+ TNF-α+, as seen in rectal mucosa (Fig. 2B).
Individuals with HLA-DRB1*13 and HLA-DQB1*06 have stronger, more polyfunctional mucosal CD4+ T-cell responses than those without the haplotype.It was interesting to note that some elite controllers had unusually high-magnitude CD4+ T-cell responses in rectal mucosa (Fig. 1A). We therefore wished to determine whether there was an association between these high responses and specific class II HLA alleles. Since a previous study had found that the haplotype HLA-DRB1*13/HLA-DQB1*06 was enriched in a cohort of long-term nonprogressors (LTNP) and that p24 lymphoproliferative responses in these individuals were more robust than in those without the haplotype (36), we initially focused on the role of this specific haplotype.
In our cohort, HLA-DRB1*13 was found only in controllers (n = 7); likewise, 16 out of 17 individuals with HLA-DQB1*06 were controllers (Fig. 3; also see Table S1 in the supplemental material). Six controllers (21%) had the combined haplotype of HLA-DRB1*13/HLA-DQB1*06, 10 controllers (36%) had the HLA-DQB1*06 allele alone, 1 controller (4%) had the HLA-DRB1*13 allele alone, and 11 controllers (39%) lacked both of these alleles. Noncontrollers in our study did not possess HLA-DRB1*13, and only one had HLA-DQB1*06. Controllers possessed HLA-DRB1*13 and/or HLA-DQB1*06 more frequently than did noncontrollers (Fig. 3) (Fisher exact test, P = 0.008). Mucosal CD4+ T cells in controllers with one or both of these class II alleles were found to have high-magnitude responses compared to those in either controllers or noncontrollers lacking these alleles (Fig. 4) (3.8% versus 2.9% [P = 0.034] or 2.1% [P = 0.048], respectively).
View larger version:FIG. 3. HLA class II alleles in the study cohort. Frequencies of controllers and noncontrollers in the current study who have HLA-DRB1*13 alone, HLA-DQB1*06 alone, the combined HLA-DRB1*13/HLA-DQB1*06 haplotype, and other class II HLA alleles not previously stated.
View larger version:FIG. 4. Total HIV Gag-specific CD4+ responses in the rectal mucosa of individuals with or without HLA-DRB1*13 and/or HLA-DQB1*06. Percentages of CD4+ T cells responding in any way (IFN-γ, IL-2, MIP-1β, and/or TNF-α) to HIV Gag stimulation in controllers with or without HLA-DRB1*13 and/or HLA-DQB1*06 and in noncontrollers lacking these alleles. (Too few noncontrollers possessed either of these alleles for a meaningful comparison.) Horizontal bars represent the median response in each group. *, P < 0.05.
Controllers with both alleles also had highly polyfunctional mucosal CD4+ T-cell responses (Fig. 5). Mucosal CD4+ T cells from controllers with the HLA-DRB1*13/HLA-DQB1*06 haplotype had significantly higher frequencies of 3-function cells (IFN-γ+ MIP-1β+ TNF-α+) than those from controllers with only the HLA-DQB1*06 allele or other class II HLA alleles (Fig. 5) (P < 0.05). There was also a trend toward higher percentages of 4-function (IFN-γ+ IL-2+ MIP-1β+ TNF-α+), 3-function (IL-2+ MIP-1β+ TNF-α+), and 2-function (IL-2+ TNF-α+) cells in the rectal mucosa of the controllers who had HLA-DRB1*13/HLA-DQB1*06 than in that of controllers without the haplotype. Controllers that had only HLA-DQB1*06 exhibited less complex responses, similar to those of subjects that had neither HLA-DRB1*13 nor HLA-DQB1*06 (Fig. 5). While there was only one controller who possessed HLA-DRB1*13 in the absence of HLA-DQB1*06, this individual had the most polyfunctional mucosal CD4+ T-cell response, with 61% of the total HIV Gag-specific CD4+ T-cell response consisting of 4 or 3 factors (data not shown).
View larger version:FIG. 5. Polyfunctional HIV Gag-specific CD4+ T-cell responses in the rectal mucosa of controllers with or without HLA-DRB1*13 and/or HLA-DQB1*06. Colors in the pie charts represent 4-function cells (black), 3-function cells (purple), 2-function cells (dark blue), and 1-function cells (light blue). Values in the pie charts are the median total CD4+ T-cell response within each group. The bar charts below show the responses in controllers with HLA-DQB1*06 alone, with the HLA-DRB1*13/HLA-DQB1*06 haplotype, or without either allele, broken down into individual functional categories using interquartile ranges. The blue asterisk below the bar chart shows a significant difference (P < 0.05) between those with the HLA-DRB1*13/HLA-DQB1*06 haplotype and those with HLA-DQB1*06 alone.
CD4+ T-cell response mapping from four controllers with HLA-DRB1*13 and/or HLA-DQB1*06.CD4+ T cells from individuals with high-magnitude total CD4+ T-cell responses in either blood or mucosa and possessing HLA-DRB1*13 and/or HLA-DQB1*06 (Fig. 4) were polyclonally expanded, and their responses to HIV Gag, Env, and Nef peptides were mapped using IFN-γ ELISpot. Four controllers had measurable responses to HIV Gag and Nef peptides (Table 1). No CD4+ T-cell responses to HIV Env peptides were detected in either blood or rectal mucosa. All four subjects responded to Gag p24, amino acids (aa) 295 to 309 (DYVDRFYKTLRAEQA), an immunodominant peptide also described by Malhotra et al. (36). Three of four subjects also had robust responses to a Gag peptide that encompasses the protease cleavage site between the p7 nucleocapsid and the p1 linker region (aa 429 to 447, RQANFLGKIQPSHKGRPGN). There were fewer CD4+ T-cell responses to HIV Nef in these individuals, and these tended to be of lower magnitude than the majority of HIV Gag responses (Table 1). While these subjects possess the HLA-DRB1*13 and/or HLA-DQB1*06 allele, the actual major histocompatibility complex (MHC) restrictions of these responses are currently unknown and will be addressed in future studies.
View this table:TABLE 1. CD4+ T-cell epitope mapping in four controllers with the HLA-DRB1*13 and/or HLA-DQB1*06 allele
It should be noted that some low-frequency T-cell populations may fail to expand in response to polyclonal stimulation, leading to underestimation of antigen-specific response frequency and/or breadth (4, 25, 26). In addition, the use of clade B consensus peptides in this study means that responses to autologous viral sequences were not measured. Nevertheless, our previous studies as well as those of other groups suggest that this approach can reliably detect and map most immunodominant and many subdominant T-cell responses (4, 17, 25, 26).
Polyfunctional CD4+ T-cell responses correlate with the magnitude of the total CD8+ T-cell response in rectal mucosa.There was a positive correlation between the magnitude of the total CD4+ T-cell response and the total CD8+ T-cell response in rectal mucosa (Fig. 6 A) (Spearman's r = 0.61, P < 0.0001). There was also a significant positive correlation between polyfunctional CD4+ T-cell responses (3 or 4 functions) and total CD8+ T-cell responses (Fig. 6B) (Spearman's r = 0.43, P = 0.005), and there was a strong trend toward a positive correlation between polyfunctional CD4+ T-cell and polyfunctional CD8+ T-cell responses (3, 4, or 5 functions) (Fig. 6C) (Spearman's r = 0.30, P = 0.053). One outlier, a controller, had no polyfunctional CD4+ T-cell response and yet had the highest polyfunctional CD8+ T-cell response (Fig. 6C). So, while in general, stronger and more polyfunctional CD4+ T-cell responses correlated with stronger and more polyfunctional CD8+ T-cell responses, this was not true for every individual.
View larger version:FIG. 6. Polyfunctional mucosal CD4+ T-cell responses correlate with the magnitude and polyfunctionality of the CD8+ T-cell response. Relationships between the magnitude of the total CD4+ T-cell response and the magnitude of the total CD8+ T-cell response (A), the frequency of the polyfunctional CD4+ T cells and the magnitude of the total CD8+ T-cell response (B), and the frequency of the polyfunctional CD4+ T cells and the frequency of the polyfunctional CD8+ T cells (C) in rectal mucosa of controllers and noncontrollers. Polyfunctional CD4+ T cells are defined as exhibiting 3 or 4 functional responses; polyfunctional CD8+ T cells are those which express 3, 4, or 5 functions. Linear-regression lines and Spearman's r correlations were calculated using the combined controller and noncontroller data set.
Previous SectionNext SectionDISCUSSIONWhile many studies have described the complex functional responses of HIV-specific CD8+ T cells, fewer reports have focused on HIV-specific CD4+ T-cell responses. Of those studies that have focused on HIV-specific CD4+ T cells, to our knowledge, all have investigated peripheral blood rather than mucosal CD4+ T-cell responses. Here we present HIV-specific CD4+ T-cell functional responses in both peripheral blood and rectal mucosa of controllers, noncontrollers, and individuals on HAART. Our findings show that HIV controllers have highly polyfunctional HIV-specific mucosal CD4+ T-cell responses compared to noncontrollers. When considered individually, levels of Gag-specific IFN-γ and MIP-1β production in the mucosa were higher in controllers than noncontrollers (Fig. 1B and C). Responses in blood generally did not reveal the same magnitude or complexity seen in the gut mucosa. The one consistent exception to these trends was the finding that, in peripheral blood, CD4+ T cells from controllers generally produced more IL-2 and TNF-α than CD4+ T cells from noncontrollers (Fig. 1E and F), as previously reported by Emu et al. (15) and Harari et al. (21, 22). Additionally, controllers had stronger and more complex HIV-specific CD4+ T-cell responses than HAART-suppressed individuals with a similarly low antigen burden, suggesting that the augmented CD4+ T-cell responses in HIV controllers are not simply a consequence of low viral load.
While the total HIV-specific CD4+ T-cell response magnitudes were similar for many controllers and noncontrollers, we noticed that some individuals who maintained viral control had unusually strong mucosal HIV Gag-specific responses, as high as 13% of the rectal CD4+ T-cell population. We hypothesized that individuals with these high responses might be enriched for certain class II HLA alleles. Indeed, controllers with HLA-DRB1*13 and/or HLA-DQB1*06, class II HLA alleles previously associated with HIV control and long-term nonprogression (10, 31, 36, 55), had the strongest mucosal CD4+ T-cell responses in our cohort. Moreover, subjects with the combined haplotype had exceptionally polyfunctional Gag-specific CD4+ T-cell responses in rectal mucosa.
When CD4+ T-cell responses were analyzed with respect to paired CD8+ T-cell responses, there was a direct correlation between polyfunctional CD4+ T-cell responses and both the magnitude and the quality of CD8+ T-cell responses in rectal mucosa. These findings were consistent with prior studies highlighting the importance of CD4+ T-cell help in maintaining functional CD8+ T-cell populations during chronic viral infection (3, 28, 29, 38, 58). It has been shown that individuals with strong Gag-specific CD8+ T-cell responses in conjunction with strong CD4+ lymphoproliferative responses are better able to control HIV-1 viremia (28). Nevertheless, some controllers in our cohort appeared to lack polyfunctional CD4+ T-cell responses and yet had strong, polyfunctional CD8+ T-cell responses. This suggests that high-quality, robust CD4+ T-cell help contributes to, but may not be essential for, robust and polyfunctional CD8+ T-cell responses. It is likely that non-T-cell-based mechanisms of control may be a factor in those controllers who lack strong, polyfunctional CD4+ and CD8+ T-cell responses, as suggested by Deeks and Walker (12). In addition, we cannot rule out the possibility that low-level ongoing viral replication in the gut mucosa of controllers could drive the expansion of both HIV-specific CD4+ and CD8+ T cells. Therefore, CD4+ and CD8+ T-cell responses could be related without being causally associated.
Although a role for MHC class I alleles in HIV control and long-term nonprogression has been unequivocally established, there is as yet no consensus on the role of specific MHC class II alleles in long-term HIV control in humans. Several previous reports have suggested an association of HLA-DRB1*13 and/or HLA-DQB1*06 with slow disease progression (10, 31, 36, 55). In a study by Malhotra et al., both long-term nonprogressors and individuals receiving antiretroviral therapy who had the HLA-DRB1*13/HLA-DQB1*06 haplotype revealed stronger Gag-specific proliferative responses and IFN-γ secretion than individuals lacking this haplotype (36). Additionally, simian immunodeficiency virus (SIV) controllers that maintain stable viral loads of <1,000 copies/ml are more likely than noncontrollers to possess the MHC class II alleles Mamu-DRB1*1003 and Mamu-DRB1*0306 (19).
It should be noted that in our study all of the subjects with the HLA-DRB1*13/HLA-DQB1*06 haplotype also had class I alleles associated with control (five with HLA-B57 and one with HLA-B81) (16). Accordingly, this study was not powered to distinguish the effects of class I versus class II alleles. However, in the Malhotra study, none of the subjects with the HLA-DRB1*13 allele had “protective” class I alleles (36). Large-scale genetic studies will be required to address the relative contributions of class I and class II alleles to HIV control (27). It is also possible that HLA-DRB1*13/HLA-DQB1*06 are in linkage disequilibrium with another factor that strongly contributes to immune control, thereby indirectly leading to relative preservation of mucosal CD4+ T cells and maintenance of strong HIV-specific CD4+ T-cell responses. This hypothesis may be supported by the observation that the HLA-DRB1*1301 and/or -1302 allele has also been associated with protection against persistent hepatitis B virus (HBV) infection, cervical carcinoma related to human papillomavirus (HPV) infection, and Plasmodium falciparum malaria (5, 24, 53).
An intriguing finding of the Malhotra study (36) was that HLA-DRB1*13-restricted CD4+ T-cell clones recognized and bound with high affinity the Gag epitopes EIYKRWIILG (EG10, p24 aa 260 to 269) and DYVDRFYKTLRAEQA (DA15, p24 aa 295 to 309), which immediately precede and partially overlap immunodominant CD8+ T-cell epitopes restricted by HLA-B27 (KK10, KRWIILGLNK, p24 aa 263 to 272) and HLA-B57 (QW9, QASQEVKNW, p24 aa 308 to 316) (18, 20). Gag p24 is highly conserved among HIV-1 subtypes, and analogous regions are found in the Gag proteins of SIV and feline immunodeficiency virus (FIV) (35, 36, 41, 42). Mutations in these regions often come at a fitness cost to the virus (40, 48) and generally involve compensatory mutations upstream of an epitope (8, 37, 47). Another recent report showed that the CD4+ T cells of controllers that responded to the Gag peptide FRDYVDRFYKTLRAEQASQE (p24 aa 293 to 312), which overlaps with DA15, had both high functional avidity and diverse TCR repertoires (54). Highly avid HIV-specific CD4+ T-cell responses directed at conserved regions within Gag may persist longer to help maintain a healthy CD8+ T-cell population, thereby aiding in the maintenance of viral suppression.
In the current study, we found that controllers with HLA-DRB1*13 and/or HLA-DQB1*06 had strong responses to the DA15 peptide; however, 3 out of 4 of these individuals had HLA-DQB1*06 alone, suggesting that these responses may not be restricted solely by HLA-DRB1*13. This would not be surprising, since class II MHC alleles are known to be highly promiscuous, and in particular the Gag peptide spanning residues 251 to 270 was previously noted to bind 10 of 12 of the most common HLA-DR alleles worldwide (30, 36, 52). Determining the class II MHC restriction of the DA15 peptide in these individuals is the focus of ongoing studies. We also observed Nef-specific responses in these four controllers, consistent with previous reports showing the immunodominance of HIV Gag and Nef among HIV-specific CD4+ T-cell responses (30).
Our data, taken together with previous findings (36), support the hypothesis that CD4+ T-cell responses act in concert with CD8+ T-cell responses as immune correlates of HIV control. High-magnitude, polyfunctional HIV-specific CD4+ T cells that reside in the rectal mucosa appear to be a strong and consistent predictor of long-term control of HIV replication in the absence of therapy. These populations are consistently more abundant in controllers than in individuals on long-term antiretroviral therapy. Since the levels of viremia are comparable in these two groups (23), it is unlikely that the observed increase in HIV-specific CD4+ T-cell responses is simply a consequence of virus control. Moreover, the finding that among HIV controllers there is an association between the presence of certain class II HLA alleles and strong CD4+ T-cell responses argues for an active role of these cells in contributing to virus control. Collectively, these data suggest that vaccine and therapeutic strategies aimed at expanding these mucosal T-cell responses may prove beneficial.
Previous SectionNext SectionACKNOWLEDGMENTSWe thank Rebecca Hoh (University of California—San Francisco) and Melissa Schreiber (University of California—Davis) for assistance with patient recruitment and enrollment. We thank the study volunteers for their contributions to this work.
This study was supported by the following sources: the National Institutes of Health (grant AI057020 to B.L.S.; grants AI069994, AI76174 and AI087145 to S.G.D.; grant AI065244 to P.W.H.; grant AI027763, the University of California, San Francisco Center for AIDS Research; grant UL1 RR024131, the University of California Clinical and Translational Science Institute); the California HIV/AIDS Research Program (grant CH05-D-606 to R.B.P.); the American Foundation for AIDS Research (grant 106710-40-RGRL to S.G.D.). This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program (grant C06 RR-12088-01) from the National Center for Research Resources, National Institutes of Health. The LSR-II violet laser was upgraded with funding from the James B. Pendleton Charitable Trust.
Previous SectionNext SectionFOOTNOTES
The study of individuals who are able to achieve durable control over human immunodeficiency virus (HIV) replication in the absence of antiretroviral therapy has become increasingly important in light of recent vaccine trials which have failed to induce protective immunity. Understanding the correlates of protection in these HIV “controllers” would aid in both the rational design of potential vaccine candidates and the testing of their efficacy.
Cell-mediated immune responses, in particular HIV-specific CD8+ T-cell responses, have been shown to be critical in decreasing the initial viremia after acute infection and determining the chronic infection set point (32, 33, 43). These responses have proven to be “too little and too late” to prevent the establishment of chronic infection; however, studies of HIV-specific CD8+ T-cell responses in controllers suggest that strong, polyfunctional responses may be important in long-term virologic control (1, 6, 11, 15, 16, 44). In addition, the association of controller status with class I HLA alleles, predominantly HLA-B27 and HLA-B57, has been well documented in multiple cohorts (2, 9, 13, 39).
The maintenance of robust HIV-specific CD4+ T-cell responses may also favor long-term control of HIV replication in untreated persons. One of the primary roles of CD4+ T cells is to provide “help” to CD8+ T cells. Many studies have shown that proper functioning of CD4+ T cells is necessary for high-quality antigen-specific CD8+ T-cell responses (3, 28, 29, 38, 58). HIV-specific CD4+ T cells in the peripheral blood of long-term nonprogressors (LTNP) have been shown to be polyfunctional, producing both gamma interferon (IFN-γ) and interleukin-2 (IL-2), whereas those from progressors tend to be monofunctional, secreting only IFN-γ (7, 14, 21, 22, 44). Additionally, the ability of CD4+ T cells to proliferate appears to be preserved in controllers (46, 56). The maturation status of CD4+ T cells that function in the context of HIV infection may also be important, as individuals who are able to preserve central memory T cells and sustain an activated effector memory CD4+ T-cell population are better able to suppress viral replication (45). Much less is known about the association of class II HLA alleles and controller status; however, a few studies have cited a relationship between HLA-DRB1*13 and/or HLA-DQB1*06 and HIV control (10, 31, 36, 55).
Previously, we have shown that CD8+ T cells from the rectal mucosa of controllers with protective class I alleles (HLA-B13, -B27, -B57, -B58, and -B81) are highly polyfunctional compared to CD8+ T cells from either controllers or noncontrollers (NC) lacking these alleles (16). Additionally, we found that mucosal CD8+ T-cell responses from individuals who had protective class I alleles in combination with the class II alleles HLA-DRB1*13 and/or HLA-DQB1*06 were of greater magnitude than mucosal CD8+ T-cell responses from those who had protective class I alleles alone (16). Therefore, we wanted to specifically examine HIV-specific mucosal CD4+ T-cell responses among controllers with and without these potentially protective class II HLA alleles.
Our hypothesis was that controllers, particularly those who possessed HLA-DRB1*13 and/or HLA-DQB1*06, would have more robust and polyfunctional HIV-specific CD4+ T-cell responses than noncontrollers or subjects on highly active antiretroviral therapy (HAART) and that these responses would correlate with strong CD8+ T-cell responses in the same individuals. We found that, indeed, controllers generally had stronger HIV-specific CD4+ T-cell responses than other groups in rectal mucosa and that, among controllers, those with the haplotype HLA-DRB1*13/HLA-DQB1*06 had particularly high percentages of polyfunctional HIV-specific CD4+ T cells in rectal mucosa. Furthermore, the proportion of polyfunctional mucosal CD4+ T cells directly correlated with the total magnitude of the mucosal CD8+ T-cell response. These data collectively suggest that the preservation or expansion of both HIV-specific CD4+ and CD8+ T cells may be an important mechanism whereby certain controllers maintain durable control of HIV replication.
Previous SectionNext SectionMATERIALS AND METHODSSubjects and tissue collection.Subjects were recruited through ongoing studies of chronic HIV infection at San Francisco General Hospital and the Center for AIDS Research, Education, and Services (CARES) Clinic in Sacramento, CA, and have been described previously (16). Subjects were classified in one of five categories based on viral load (VL) measurements and treatment status, as described by Deeks and Walker (12): elite controllers (EC; VL of <75 copies/ml, off therapy, n = 17), viremic controllers (VC; VL of <2,000 copies/ml, off therapy, n = 11), noncontrollers (NC; VL of >10,000 copies, off therapy, n = 14), highly active antiretroviral therapy (HAART)-suppressed subjects (VL of <75 copies, on HAART, n = 10), and HIV-negative controls (n = 8). Written informed consent for phlebotomy and flexible sigmoidoscopy was obtained from all subjects in accordance with the Declaration of Helsinki, and study protocols were approved by the Institutional Review Board, University of California—Davis, and the Committee on Human Subjects Research, University of California—San Francisco.
Approximately 20 ml of blood was obtained by sterile venipuncture and collected into tubes containing EDTA. Rectal biopsy tissue (20 to 25 pieces) was obtained by flexible sigmoidoscopy at 10 cm from the anal verge using a sigmoidoscope equipped with a biopsy channel and single-use biopsy forceps. This procedure has been well documented to cause only minimal discomfort and provide enough cells to perform cellular immunology assays (mean of 10 × 106 cells) (11, 16, 49, 51). Tissue was placed in RPMI 1640 supplemented with 15% fetal calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and l-glutamine (2 nM), herein referred to as R15. Specimens were immediately transported to the University of California—Davis for same-day processing.
Peripheral blood and rectal biopsy tissue processing.Peripheral blood was layered onto a Ficoll-Hypaque (Pfizer, New York, NY) density gradient to isolate mononuclear cells (PBMC). Rectal biopsy tissue was processed according to a previously published protocol designed to maximize viable lymphocyte yield (11, 16, 49, 51). Briefly, biopsy tissue was subjected to three rounds of collagenase type II digestion (0.5 mg/ml; Sigma-Aldrich, St. Louis, MO), followed by mechanical disruption using an 18-gauge blunt-end needle and passage through a 70-μm cell strainer. Cells were then washed in R15 and layered on a 35%/65% Percoll gradient (Sigma-Aldrich). Mononuclear cells were harvested from both the medium-35% Percoll interface and the 35%/65% Percoll interface to maximize yield. PBMC and rectal mononuclear cells (RMC) were rested overnight at 37°C and 5% CO2, and piperacillin-tazobactam (Zosyn, 0.5 mg/ml; Wyeth-Ayerst, Princeton, NJ) was added to RMC cultures to prevent bacterial growth.
HLA class II typing.Genomic DNA was isolated from 5 × 106 to 8 × 106 PBMC by using a QIAamp DNA blood minikit (Qiagen, Valencia, CA) and quantified by spectrophotometer. Low-resolution HLA-DR and HLA-DQ typing was performed using PCR and a DR/DQ 2T locus SSP (sequence-specific primer) Unitray kit (Invitrogen, Carlsbad, CA). PCR products were resolved by electrophoresis on a 2% agarose gel, photographed, and analyzed using UniMatch Plus software (Invitrogen).
Antibodies and peptide pools.Fluorochrome-labeled monoclonal antibodies to CD3 (clone UCHT-1), CD107a (clone H4A3), gamma interferon (IFN-γ; clone B27), macrophage inflammatory protein 1β (MIP-1β; clone D21-1351), tumor necrosis factor alpha (TNF-α; clone MAb11), and interleukin-2 (IL-2; clone 5344.111) and unlabeled antibodies to CD28 (clone L293) and CD49d (clone L25) were purchased from BD Biosciences (San Jose, CA). Fluorochrome-labeled antibodies to CD4 (clone SFCI12T4D11) and CD8 (clone SK1) were purchased from Beckman Coulter (Fullerton, CA) and Invitrogen, respectively. All antibodies were titrated to determine optimum concentrations for assay conditions (data not shown). HIV Gag (p55, HXB2 sequence) peptide pools containing 15-mer peptides overlapping by 11 amino acids (aa) were purchased from BD Biosciences.
Intracellular cytokine staining and flow cytometry.Intracellular cytokine staining of newly isolated PBMC or RMC without prior expansion was performed as previously described (16). Briefly, 1 × 106 to 2 × 106 PBMC or RMC were stimulated for 5 h with the HIV Gag p55 pool (3.5 μg/ml) in the presence of anti-CD28 (2.5 μg/ml) and anti-CD49d (5 μg/ml) costimulatory antibodies, anti-CD107a, Golgi Stop (1 μM; BD Biosciences), and brefeldin A (5 μg/ml; Sigma-Aldrich). Dimethyl sulfoxide (DMSO) and Staphylococcus enterotoxin B (SEB; 5 μg/ml) served as negative and positive controls, respectively. Cells were then stained with antibodies to surface antigens CD4 and CD8 and with 7-amino-actinomycin D (7-AAD; BD Biosciences) to label dead cells. Thereafter, all buffers and reagents contained actinomycin D to saturate sites with the potential to bind 7-AAD. Cells were fixed in 4% paraformaldehyde and then permeabilized using FACS Perm 2 (BD Biosciences). Intracellular cytokine staining for CD3, IFN-γ, IL-2, MIP-1β, and TNF-α followed permeabilization. Finally, the cells were placed in 1% paraformaldehyde for at least 1 h, but no longer than 24 h, before being read on an LSRII flow cytometer equipped with a 405-, 488-, and 643-nm laser (BD Biosciences).
Polyclonal CD4+ T-cell expansion of peripheral blood and rectal mononuclear cells.Polyclonal expansion was performed in order to obtain sufficient CD4+ T cells for epitope mapping using the enzyme-linked immunospot (ELISpot) method (47). This approach has been shown to expand T cells nonspecifically without significantly altering T-cell receptor (TCR) clonotypes, patterns of epitope recognition, or the ability of T cells to respond to peptide stimulation by secreting IFN-γ (4, 25, 26). It should be noted that, in the present study, polyclonal expansion was utilized only to generate sufficient cells to map the peptide specificity of responses that had previously been identified by intracellular cytokine staining of fresh PBMC and RMC. One million PBMC or RMC were placed in 2 ml R15 in a 24-well plate with 1 μg/ml CD3/CD8-bispecific antibody (generously provided by Johnson Wong, Harvard University), which preferentially expands CD4+ T cells while promoting apoptosis in CD8+ T cells (57). PBMC and RMC cultures were supplemented with recombinant IL-2 (rIL-2) (50 U/ml; R&D Systems, Minneapolis, MN).
When initial attempts to culture RMC (but not PBMC) gave a low success rate, we supplemented cultures with 1 ng/ml human recombinant IL-7 (R&D Systems, Minneapolis, MN), based on a previously published protocol. This cytokine promotes survival of memory T cells, in part by inactivating proapoptotic pathways (34). Amphotericin B (1.25 μg/ml; MP Biomedicals, Solon, OH) and piperacillin-tazobactam (Zosyn, 0.5 μg/ml; Wyeth-Ayerst, Princeton, NJ) were added to prevent fungal and bacterial growth, respectively. After the first week, IL-2 (and IL-7 for RMC cultures) was added twice weekly, as were additional amphotericin B and piperacillin-tazobactam, to compensate for any added R15 volume during the expansion. Two to 3 days following the initial CD3/CD8 antibody stimulation, 2 × 106 irradiated heterologous PBMC (5,000 rad) from a seronegative donor were added to cultures. Cultures were expanded and maintained for 4 to 8 weeks. After 3 weeks of culture, cells were restimulated with 0.1 μg/ml 12F6 anti-CD3 antibody (also provided by Johnson Wong, Harvard University).
CD4+ T-cell response mapping.By 4 to 5 weeks, polyclonally expanded cultures were composed of primarily CD4+ T cells; however, approximately half of the cultures included 20 to 40% CD8+ T cells. Therefore, CD8+ T cells were depleted from cultures by using magnetic bead separation prior to epitope mapping. Anti-CD8 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) were used to label CD8+ T cells, which were then removed from the CD4+ T cells by using a MACS system (Miltenyi Biotec) with an LD column.
CD4+ T-cell response mapping was performed using an IFN-γ ELISpot assay as previously described for CD8+ T-cell response mapping (50). Briefly, pools of HIV HXB2 consensus clade B Gag, Env, and Nef peptides (15-mer peptides overlapping by 11; NIH AIDS Research and Reference Reagent Program, Rockville, MD) were created so that each peptide was contained in exactly two pools. This allowed the creation of a peptide matrix for screening HIV peptides. Peptides common to pools positive for a CD4+ T-cell response (>50 spot-forming cells [SFC]/million after the subtraction of background) were then tested individually using a standard IFN-γ ELISpot assay.
Data analysis.Flow cytometry data were analyzed with FlowJo software (TreeStar, Ashland, OR). Boolean gate analysis allowed the separation of CD4+ T-cell responses into 16 individual combinations of the four functional parameters (IFN-γ, IL-2, MIP-1β, and TNF-α). Data generated from Boolean gate analysis were then visualized in SPICE software (v.4.2.2; Mario Roederer, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD). Standard bivariate plots were also constructed to confirm the placement of individual Boolean gates.
IFN-γ ELISpots were read on an AID plate reader (Autoimmun Diagnostika GMBH, Straßerg, Germany), and pool- or peptide-specific CD4+ T-cell responses were quantified as SFC/million after the subtraction of background. Responses of >50 SFC/million were considered positive.
Statistical analysis.CD4+ T-cell flow cytometry data were analyzed using a previously described formula which considers whether or not antigen-specific responses differ significantly from background (11, 16). This step was important for evaluating mucosal T-cell responses, as they are highly responsive to costimulation (data not shown) and can produce low levels of cytokines and chemokines in the absence of antigen-specific stimulation. The formula assumes a Poisson distribution and takes into account the actual number of collected events rather than percentages of responding cells, as the lymphocyte yield from rectal mucosa tends to be low. When a determination that antigen-specific values were significantly different from background was made, net responses were then calculated by subtracting unstimulated control values from antigen-specific values. Responses that were not determined to be significantly different from background were dropped (11, 16).
For response data, comparisons between two groups were made using a two-tailed Mann-Whitney test in GraphPad Prism software (GraphPad Software, San Diego, CA). Comparisons between polyfunctionality pie charts utilized a permutation test based on χ2 statistics, and comparisons between two individual functional categories employed a Wilcoxon rank test (SPICE Software). A two-tailed Fisher exact test was used for comparing the distributions of HLA-DRB1*13 and HLA-DQB1*06 among subject groups. Correlations between two variables were done using the Spearman correlation, and linear regression was used to graph a best-fit line to the data (GraphPad Prism Software).
Previous SectionNext SectionRESULTSBaseline characteristics.The individuals in this study have been described in an earlier report (16). Briefly, 52 HIV-positive individuals were assigned to one of four groups, as described in Materials and Methods: elite controllers, viremic controllers, noncontrollers, and HAART-suppressed patients. Eight HIV-negative subjects were included as controls.
The percentage of CD4+ T cells as a proportion of CD3+ T cells in peripheral blood and rectal mucosa was previously determined by flow cytometry (16). Elite and viremic controllers had significantly higher percentages of CD4+ T cells in rectal mucosa than noncontrollers (medians of 42.2% and 30.1% versus 15.1%, respectively; P < 0.05); rectal CD4+ percentages in controllers were comparable to those in subjects on HAART (median of 45.8%). All HIV-positive individuals had lower percentages of CD4+ T cells than HIV-negative controls (median of 73%; P < 0.01) (16).
High-magnitude HIV-specific CD4+ T-cell responses in rectal mucosa.The ability of CD4+ T cells to respond to HIV Gag stimulation by producing IFN-γ, IL-2, MIP-1β, and/or TNF-α was measured by flow cytometry using fresh PBMC and RMC without prior expansion (see reference 16 for gating strategy). CD4+ T-cell responses were generally much higher, in terms of total percentage of responding cells, in rectal mucosa than peripheral blood. This likely reflects the predominance of antigen-experienced memory cells in rectal mucosa and was previously observed for HIV-specific CD8+ T cells (16). The median total HIV-specific mucosal CD4+ T-cell response (cells positive for one or more of the above-described functions, all responses combined) was significantly higher in elite controllers than in individuals on HAART (Fig. 1 A) (3.4% versus 0.4%, respectively; P = 0.045). While some elite controllers had unusually strong Gag-specific mucosal CD4+ T-cell responses (as high as 13%), responses within the group were highly heterogeneous, and therefore the median total CD4+ T-cell responses were not significantly different between elite controllers and noncontrollers (3.4% versus 2.3%, respectively; P = 0.21).
View larger version:FIG. 1. HIV Gag-specific CD4+ T-cell responses in rectal mucosa (A to C) and peripheral blood (D to F). Panels A and D show the total percentages of CD4+ T cells capable of producing IFN-γ, IL-2, MIP-1β, and/or TNF-α in response to HIV Gag stimulation in rectal mucosa (EC versus subjects on HAART, P = 0.045) (A) and peripheral blood (D). Panels B and C show significant differences in CD4+ T-cell cytokine/chemokine expression in rectal mucosa between groups: IFN-γ (EC versus NC, P = 0.031; EC versus subjects on HAART, P = 0.035; VC versus NC, P = 0.0005; VC versus subjects on HAART, P = 0.004) (B); MIP-1β (EC versus NC, P = 0.004; EC versus subjects on HAART, P = 0.016; VC versus NC, P = 0.012) (C). Panels E and F show significant differences in CD4+ T-cell cytokine expression in peripheral blood between groups: IL-2 (EC versus NC, P = 0.008; EC versus subjects on HAART, P = 0.018; VC versus NC, P = 0.003; VC versus subjects on HAART, P = 0.001) (E); TNF-α (VC versus NC, P = 0.019; VC versus subjects on HAART, P = 0.006) (F). Horizontal bars represent the median response for each group. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
While predominantly tissue-specific differences (i.e., between peripheral blood and rectal mucosa) were found in the overall magnitude of HIV-specific CD4+ T-cell responses, differences were found in CD4+ T-cell responses between subject groups when assessed for individual functions. Elite controllers and viremic controllers had higher frequencies of mucosal CD4+ T cells producing IFN-γ (Fig. 1B) and MIP-1β (Fig. 1C) than either noncontrollers (NC) or HAART-suppressed individuals.
In general, <0.5% of peripheral blood CD4+ T cells from controllers, noncontrollers, or HAART-suppressed subjects responded in any way to Gag stimulation (Fig. 1D). There was no consistent difference the in ability of mucosal CD4+ T cells to produce IL-2 or TNF-α across subject groups; however, controllers generally had higher levels of IL-2-producing cells in peripheral blood (Fig. 1E), as previously described by our group and others (7, 14, 21, 44), as well as higher levels of TNF-α (Fig. 1F).
HIV controllers have complex Gag-specific mucosal CD4+ T-cell responses.Boolean gate analysis was used to assess the complexity of the HIV-specific CD4+ T-cell response by looking at all possible combinations of four functional parameters: IFN-γ, IL-2, TNF-α, and MIP-1β. Cells that simultaneously expressed three or four cytokines in response to HIV peptide stimulation were considered “polyfunctional.” As there were no significant differences in polyfunctionality between elite and viremic controllers (data not shown), the two groups were combined into a single “controller” group in order to simplify the analysis. Controllers showed a trend toward a mucosal CD4+ T-cell response that was overall more polyfunctional than that for noncontrollers, as represented by the pie charts in Fig. 2 A (P = 0.051). Broken down into the 16 separate functional categories, controllers had higher frequencies of four-function mucosal CD4+ T cells than noncontrollers and more three-function CD4+ T cells (primarily IFN-γ+ MIP-1β+ TNF-α+ and lacking IL-2) than noncontrollers or individuals on HAART (Fig. 2A) (P < 0.05). Interestingly, controllers showed high levels of MIP-1β single positive cells in rectal mucosa, while noncontrollers had high levels of TNF-α single positive cells, compared to levels in other subject groups (P < 0.05).
View larger version:FIG. 2. HIV Gag-specific CD4+ T-cell polyfunctional responses in rectal mucosa and peripheral blood across subject groups. CD4+ T-cell polyfunctional responses in the rectal mucosa (A) and peripheral blood (B) of controllers, noncontrollers, and HAART-suppressed individuals. Colors in the pie charts represent 4-function cells (black), 3-function cells (purple), 2-function cells (dark blue), and 1-function cells (light blue). Values in the pie charts are the median total CD4+ T-cell response within each subject group. The bar charts below show the responses in controllers, noncontrollers, and HAART-suppressed individuals, broken down into individual functional categories using interquartile ranges. Asterisks below the bar charts show significant differences (P < 0.05; Wilcoxon rank test) between controllers and noncontrollers (red) or subjects on HAART (green).
The response complexity in peripheral blood of HIV controllers appeared to be more polyfunctional than that in noncontrollers or HAART-suppressed subjects, but these differences did not reach statistical significance (Fig. 2B). Similar to responses seen in rectal mucosa, controllers had higher percentages of Gag-specific CD4+ T cells capable of three or four functions than noncontrollers (P < 0.05 for peripheral blood); however, the dominant three-function category (in terms of median response) in blood was IFN-γ+ IL-2+ TNF-α+, rather than IFN-γ+ MIP-1β+ TNF-α+, as seen in rectal mucosa (Fig. 2B).
Individuals with HLA-DRB1*13 and HLA-DQB1*06 have stronger, more polyfunctional mucosal CD4+ T-cell responses than those without the haplotype.It was interesting to note that some elite controllers had unusually high-magnitude CD4+ T-cell responses in rectal mucosa (Fig. 1A). We therefore wished to determine whether there was an association between these high responses and specific class II HLA alleles. Since a previous study had found that the haplotype HLA-DRB1*13/HLA-DQB1*06 was enriched in a cohort of long-term nonprogressors (LTNP) and that p24 lymphoproliferative responses in these individuals were more robust than in those without the haplotype (36), we initially focused on the role of this specific haplotype.
In our cohort, HLA-DRB1*13 was found only in controllers (n = 7); likewise, 16 out of 17 individuals with HLA-DQB1*06 were controllers (Fig. 3; also see Table S1 in the supplemental material). Six controllers (21%) had the combined haplotype of HLA-DRB1*13/HLA-DQB1*06, 10 controllers (36%) had the HLA-DQB1*06 allele alone, 1 controller (4%) had the HLA-DRB1*13 allele alone, and 11 controllers (39%) lacked both of these alleles. Noncontrollers in our study did not possess HLA-DRB1*13, and only one had HLA-DQB1*06. Controllers possessed HLA-DRB1*13 and/or HLA-DQB1*06 more frequently than did noncontrollers (Fig. 3) (Fisher exact test, P = 0.008). Mucosal CD4+ T cells in controllers with one or both of these class II alleles were found to have high-magnitude responses compared to those in either controllers or noncontrollers lacking these alleles (Fig. 4) (3.8% versus 2.9% [P = 0.034] or 2.1% [P = 0.048], respectively).
View larger version:FIG. 3. HLA class II alleles in the study cohort. Frequencies of controllers and noncontrollers in the current study who have HLA-DRB1*13 alone, HLA-DQB1*06 alone, the combined HLA-DRB1*13/HLA-DQB1*06 haplotype, and other class II HLA alleles not previously stated.
View larger version:FIG. 4. Total HIV Gag-specific CD4+ responses in the rectal mucosa of individuals with or without HLA-DRB1*13 and/or HLA-DQB1*06. Percentages of CD4+ T cells responding in any way (IFN-γ, IL-2, MIP-1β, and/or TNF-α) to HIV Gag stimulation in controllers with or without HLA-DRB1*13 and/or HLA-DQB1*06 and in noncontrollers lacking these alleles. (Too few noncontrollers possessed either of these alleles for a meaningful comparison.) Horizontal bars represent the median response in each group. *, P < 0.05.
Controllers with both alleles also had highly polyfunctional mucosal CD4+ T-cell responses (Fig. 5). Mucosal CD4+ T cells from controllers with the HLA-DRB1*13/HLA-DQB1*06 haplotype had significantly higher frequencies of 3-function cells (IFN-γ+ MIP-1β+ TNF-α+) than those from controllers with only the HLA-DQB1*06 allele or other class II HLA alleles (Fig. 5) (P < 0.05). There was also a trend toward higher percentages of 4-function (IFN-γ+ IL-2+ MIP-1β+ TNF-α+), 3-function (IL-2+ MIP-1β+ TNF-α+), and 2-function (IL-2+ TNF-α+) cells in the rectal mucosa of the controllers who had HLA-DRB1*13/HLA-DQB1*06 than in that of controllers without the haplotype. Controllers that had only HLA-DQB1*06 exhibited less complex responses, similar to those of subjects that had neither HLA-DRB1*13 nor HLA-DQB1*06 (Fig. 5). While there was only one controller who possessed HLA-DRB1*13 in the absence of HLA-DQB1*06, this individual had the most polyfunctional mucosal CD4+ T-cell response, with 61% of the total HIV Gag-specific CD4+ T-cell response consisting of 4 or 3 factors (data not shown).
View larger version:FIG. 5. Polyfunctional HIV Gag-specific CD4+ T-cell responses in the rectal mucosa of controllers with or without HLA-DRB1*13 and/or HLA-DQB1*06. Colors in the pie charts represent 4-function cells (black), 3-function cells (purple), 2-function cells (dark blue), and 1-function cells (light blue). Values in the pie charts are the median total CD4+ T-cell response within each group. The bar charts below show the responses in controllers with HLA-DQB1*06 alone, with the HLA-DRB1*13/HLA-DQB1*06 haplotype, or without either allele, broken down into individual functional categories using interquartile ranges. The blue asterisk below the bar chart shows a significant difference (P < 0.05) between those with the HLA-DRB1*13/HLA-DQB1*06 haplotype and those with HLA-DQB1*06 alone.
CD4+ T-cell response mapping from four controllers with HLA-DRB1*13 and/or HLA-DQB1*06.CD4+ T cells from individuals with high-magnitude total CD4+ T-cell responses in either blood or mucosa and possessing HLA-DRB1*13 and/or HLA-DQB1*06 (Fig. 4) were polyclonally expanded, and their responses to HIV Gag, Env, and Nef peptides were mapped using IFN-γ ELISpot. Four controllers had measurable responses to HIV Gag and Nef peptides (Table 1). No CD4+ T-cell responses to HIV Env peptides were detected in either blood or rectal mucosa. All four subjects responded to Gag p24, amino acids (aa) 295 to 309 (DYVDRFYKTLRAEQA), an immunodominant peptide also described by Malhotra et al. (36). Three of four subjects also had robust responses to a Gag peptide that encompasses the protease cleavage site between the p7 nucleocapsid and the p1 linker region (aa 429 to 447, RQANFLGKIQPSHKGRPGN). There were fewer CD4+ T-cell responses to HIV Nef in these individuals, and these tended to be of lower magnitude than the majority of HIV Gag responses (Table 1). While these subjects possess the HLA-DRB1*13 and/or HLA-DQB1*06 allele, the actual major histocompatibility complex (MHC) restrictions of these responses are currently unknown and will be addressed in future studies.
View this table:TABLE 1. CD4+ T-cell epitope mapping in four controllers with the HLA-DRB1*13 and/or HLA-DQB1*06 allele
It should be noted that some low-frequency T-cell populations may fail to expand in response to polyclonal stimulation, leading to underestimation of antigen-specific response frequency and/or breadth (4, 25, 26). In addition, the use of clade B consensus peptides in this study means that responses to autologous viral sequences were not measured. Nevertheless, our previous studies as well as those of other groups suggest that this approach can reliably detect and map most immunodominant and many subdominant T-cell responses (4, 17, 25, 26).
Polyfunctional CD4+ T-cell responses correlate with the magnitude of the total CD8+ T-cell response in rectal mucosa.There was a positive correlation between the magnitude of the total CD4+ T-cell response and the total CD8+ T-cell response in rectal mucosa (Fig. 6 A) (Spearman's r = 0.61, P < 0.0001). There was also a significant positive correlation between polyfunctional CD4+ T-cell responses (3 or 4 functions) and total CD8+ T-cell responses (Fig. 6B) (Spearman's r = 0.43, P = 0.005), and there was a strong trend toward a positive correlation between polyfunctional CD4+ T-cell and polyfunctional CD8+ T-cell responses (3, 4, or 5 functions) (Fig. 6C) (Spearman's r = 0.30, P = 0.053). One outlier, a controller, had no polyfunctional CD4+ T-cell response and yet had the highest polyfunctional CD8+ T-cell response (Fig. 6C). So, while in general, stronger and more polyfunctional CD4+ T-cell responses correlated with stronger and more polyfunctional CD8+ T-cell responses, this was not true for every individual.
View larger version:FIG. 6. Polyfunctional mucosal CD4+ T-cell responses correlate with the magnitude and polyfunctionality of the CD8+ T-cell response. Relationships between the magnitude of the total CD4+ T-cell response and the magnitude of the total CD8+ T-cell response (A), the frequency of the polyfunctional CD4+ T cells and the magnitude of the total CD8+ T-cell response (B), and the frequency of the polyfunctional CD4+ T cells and the frequency of the polyfunctional CD8+ T cells (C) in rectal mucosa of controllers and noncontrollers. Polyfunctional CD4+ T cells are defined as exhibiting 3 or 4 functional responses; polyfunctional CD8+ T cells are those which express 3, 4, or 5 functions. Linear-regression lines and Spearman's r correlations were calculated using the combined controller and noncontroller data set.
Previous SectionNext SectionDISCUSSIONWhile many studies have described the complex functional responses of HIV-specific CD8+ T cells, fewer reports have focused on HIV-specific CD4+ T-cell responses. Of those studies that have focused on HIV-specific CD4+ T cells, to our knowledge, all have investigated peripheral blood rather than mucosal CD4+ T-cell responses. Here we present HIV-specific CD4+ T-cell functional responses in both peripheral blood and rectal mucosa of controllers, noncontrollers, and individuals on HAART. Our findings show that HIV controllers have highly polyfunctional HIV-specific mucosal CD4+ T-cell responses compared to noncontrollers. When considered individually, levels of Gag-specific IFN-γ and MIP-1β production in the mucosa were higher in controllers than noncontrollers (Fig. 1B and C). Responses in blood generally did not reveal the same magnitude or complexity seen in the gut mucosa. The one consistent exception to these trends was the finding that, in peripheral blood, CD4+ T cells from controllers generally produced more IL-2 and TNF-α than CD4+ T cells from noncontrollers (Fig. 1E and F), as previously reported by Emu et al. (15) and Harari et al. (21, 22). Additionally, controllers had stronger and more complex HIV-specific CD4+ T-cell responses than HAART-suppressed individuals with a similarly low antigen burden, suggesting that the augmented CD4+ T-cell responses in HIV controllers are not simply a consequence of low viral load.
While the total HIV-specific CD4+ T-cell response magnitudes were similar for many controllers and noncontrollers, we noticed that some individuals who maintained viral control had unusually strong mucosal HIV Gag-specific responses, as high as 13% of the rectal CD4+ T-cell population. We hypothesized that individuals with these high responses might be enriched for certain class II HLA alleles. Indeed, controllers with HLA-DRB1*13 and/or HLA-DQB1*06, class II HLA alleles previously associated with HIV control and long-term nonprogression (10, 31, 36, 55), had the strongest mucosal CD4+ T-cell responses in our cohort. Moreover, subjects with the combined haplotype had exceptionally polyfunctional Gag-specific CD4+ T-cell responses in rectal mucosa.
When CD4+ T-cell responses were analyzed with respect to paired CD8+ T-cell responses, there was a direct correlation between polyfunctional CD4+ T-cell responses and both the magnitude and the quality of CD8+ T-cell responses in rectal mucosa. These findings were consistent with prior studies highlighting the importance of CD4+ T-cell help in maintaining functional CD8+ T-cell populations during chronic viral infection (3, 28, 29, 38, 58). It has been shown that individuals with strong Gag-specific CD8+ T-cell responses in conjunction with strong CD4+ lymphoproliferative responses are better able to control HIV-1 viremia (28). Nevertheless, some controllers in our cohort appeared to lack polyfunctional CD4+ T-cell responses and yet had strong, polyfunctional CD8+ T-cell responses. This suggests that high-quality, robust CD4+ T-cell help contributes to, but may not be essential for, robust and polyfunctional CD8+ T-cell responses. It is likely that non-T-cell-based mechanisms of control may be a factor in those controllers who lack strong, polyfunctional CD4+ and CD8+ T-cell responses, as suggested by Deeks and Walker (12). In addition, we cannot rule out the possibility that low-level ongoing viral replication in the gut mucosa of controllers could drive the expansion of both HIV-specific CD4+ and CD8+ T cells. Therefore, CD4+ and CD8+ T-cell responses could be related without being causally associated.
Although a role for MHC class I alleles in HIV control and long-term nonprogression has been unequivocally established, there is as yet no consensus on the role of specific MHC class II alleles in long-term HIV control in humans. Several previous reports have suggested an association of HLA-DRB1*13 and/or HLA-DQB1*06 with slow disease progression (10, 31, 36, 55). In a study by Malhotra et al., both long-term nonprogressors and individuals receiving antiretroviral therapy who had the HLA-DRB1*13/HLA-DQB1*06 haplotype revealed stronger Gag-specific proliferative responses and IFN-γ secretion than individuals lacking this haplotype (36). Additionally, simian immunodeficiency virus (SIV) controllers that maintain stable viral loads of <1,000 copies/ml are more likely than noncontrollers to possess the MHC class II alleles Mamu-DRB1*1003 and Mamu-DRB1*0306 (19).
It should be noted that in our study all of the subjects with the HLA-DRB1*13/HLA-DQB1*06 haplotype also had class I alleles associated with control (five with HLA-B57 and one with HLA-B81) (16). Accordingly, this study was not powered to distinguish the effects of class I versus class II alleles. However, in the Malhotra study, none of the subjects with the HLA-DRB1*13 allele had “protective” class I alleles (36). Large-scale genetic studies will be required to address the relative contributions of class I and class II alleles to HIV control (27). It is also possible that HLA-DRB1*13/HLA-DQB1*06 are in linkage disequilibrium with another factor that strongly contributes to immune control, thereby indirectly leading to relative preservation of mucosal CD4+ T cells and maintenance of strong HIV-specific CD4+ T-cell responses. This hypothesis may be supported by the observation that the HLA-DRB1*1301 and/or -1302 allele has also been associated with protection against persistent hepatitis B virus (HBV) infection, cervical carcinoma related to human papillomavirus (HPV) infection, and Plasmodium falciparum malaria (5, 24, 53).
An intriguing finding of the Malhotra study (36) was that HLA-DRB1*13-restricted CD4+ T-cell clones recognized and bound with high affinity the Gag epitopes EIYKRWIILG (EG10, p24 aa 260 to 269) and DYVDRFYKTLRAEQA (DA15, p24 aa 295 to 309), which immediately precede and partially overlap immunodominant CD8+ T-cell epitopes restricted by HLA-B27 (KK10, KRWIILGLNK, p24 aa 263 to 272) and HLA-B57 (QW9, QASQEVKNW, p24 aa 308 to 316) (18, 20). Gag p24 is highly conserved among HIV-1 subtypes, and analogous regions are found in the Gag proteins of SIV and feline immunodeficiency virus (FIV) (35, 36, 41, 42). Mutations in these regions often come at a fitness cost to the virus (40, 48) and generally involve compensatory mutations upstream of an epitope (8, 37, 47). Another recent report showed that the CD4+ T cells of controllers that responded to the Gag peptide FRDYVDRFYKTLRAEQASQE (p24 aa 293 to 312), which overlaps with DA15, had both high functional avidity and diverse TCR repertoires (54). Highly avid HIV-specific CD4+ T-cell responses directed at conserved regions within Gag may persist longer to help maintain a healthy CD8+ T-cell population, thereby aiding in the maintenance of viral suppression.
In the current study, we found that controllers with HLA-DRB1*13 and/or HLA-DQB1*06 had strong responses to the DA15 peptide; however, 3 out of 4 of these individuals had HLA-DQB1*06 alone, suggesting that these responses may not be restricted solely by HLA-DRB1*13. This would not be surprising, since class II MHC alleles are known to be highly promiscuous, and in particular the Gag peptide spanning residues 251 to 270 was previously noted to bind 10 of 12 of the most common HLA-DR alleles worldwide (30, 36, 52). Determining the class II MHC restriction of the DA15 peptide in these individuals is the focus of ongoing studies. We also observed Nef-specific responses in these four controllers, consistent with previous reports showing the immunodominance of HIV Gag and Nef among HIV-specific CD4+ T-cell responses (30).
Our data, taken together with previous findings (36), support the hypothesis that CD4+ T-cell responses act in concert with CD8+ T-cell responses as immune correlates of HIV control. High-magnitude, polyfunctional HIV-specific CD4+ T cells that reside in the rectal mucosa appear to be a strong and consistent predictor of long-term control of HIV replication in the absence of therapy. These populations are consistently more abundant in controllers than in individuals on long-term antiretroviral therapy. Since the levels of viremia are comparable in these two groups (23), it is unlikely that the observed increase in HIV-specific CD4+ T-cell responses is simply a consequence of virus control. Moreover, the finding that among HIV controllers there is an association between the presence of certain class II HLA alleles and strong CD4+ T-cell responses argues for an active role of these cells in contributing to virus control. Collectively, these data suggest that vaccine and therapeutic strategies aimed at expanding these mucosal T-cell responses may prove beneficial.
Previous SectionNext SectionACKNOWLEDGMENTSWe thank Rebecca Hoh (University of California—San Francisco) and Melissa Schreiber (University of California—Davis) for assistance with patient recruitment and enrollment. We thank the study volunteers for their contributions to this work.
This study was supported by the following sources: the National Institutes of Health (grant AI057020 to B.L.S.; grants AI069994, AI76174 and AI087145 to S.G.D.; grant AI065244 to P.W.H.; grant AI027763, the University of California, San Francisco Center for AIDS Research; grant UL1 RR024131, the University of California Clinical and Translational Science Institute); the California HIV/AIDS Research Program (grant CH05-D-606 to R.B.P.); the American Foundation for AIDS Research (grant 106710-40-RGRL to S.G.D.). This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program (grant C06 RR-12088-01) from the National Center for Research Resources, National Institutes of Health. The LSR-II violet laser was upgraded with funding from the James B. Pendleton Charitable Trust.
Previous SectionNext SectionFOOTNOTES
- Received 5 May 2010.
- Accepted 3 August 2010.
- ↵*Corresponding author. Mailing address: Dept. of Medical Microbiology and Immunology, School of Medicine, University of California, 3327 Tupper Hall, 1 Shields Ave., Davis, CA 95616. Phone: (530) 752-6785. Fax: (530) 752-8692. E-mail: [email protected]
- ↵▿ Published ahead of print on 18 August 2010.
- ↵† Supplemental material for this article may be found at http://jvi.asm.org/.
- ↵ Almeida, J. R., D. A. Price, L. Papagno, Z. A. Arkoub, D. Sauce, E. Bornstein, T. E. Asher, A. Samri, A. Schnuriger, I. Theodorou, D. Costagliola, C. Rouzioux, H. Agut, A. G. Marcelin, D. Douek, B. Autran, and V. Appay. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473-2485. Abstract/FREE Full Text
- ↵ Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3:e403. CrossRefMedline
- ↵ Altfeld, M., and E. S. Rosenberg. 2000. The role of CD4(+) T helper cells in the cytotoxic T lymphocyte response to HIV-1. Curr. Opin. Immunol. 12:375-380. CrossRefMedline
- ↵ Altfeld, M., J. van Lunzen, N. Frahm, X. G. Yu, C. Schneider, R. L. Eldridge, M. E. Feeney, D. Meyer-Olson, H. J. Stellbrink, and B. D. Walker. 2002. Expansion of pre-existing, lymph node-localized CD8+ T cells during supervised treatment interruptions in chronic HIV-1 infection. J. Clin. Invest. 109:837-843. CrossRefMedline
- ↵ Apple, R. J., H. A. Erlich, W. Klitz, M. M. Manos, T. M. Becker, and C. M. Wheeler. 1994. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat. Genet. 6:157-162. CrossRefMedline
- ↵ Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781-4789. Abstract/FREE Full Text
- ↵ Boaz, M. J., A. Waters, S. Murad, P. J. Easterbrook, and A. Vyakarnam. 2002. Presence of HIV-1 Gag-specific IFN-gamma+IL-2+ and CD28+IL-2+ CD4 T cell responses is associated with nonprogression in HIV-1 infection. J. Immunol. 169:6376-6385. Abstract/FREE Full Text
- ↵ Brockman, M. A., A. Schneidewind, M. Lahaie, A. Schmidt, T. Miura, I. Desouza, F. Ryvkin, C. A. Derdeyn, S. Allen, E. Hunter, J. Mulenga, P. A. Goepfert, B. D. Walker, and T. M. Allen. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608-12618. Abstract/FREE Full Text
- ↵ Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535-551. CrossRefMedline
- ↵ Chen, Y., R. Winchester, B. Korber, J. Gagliano, Y. Bryson, C. Hutto, N. Martin, G. McSherry, A. Petru, D. Wara, and A. Ammann. 1997. Influence of HLA alleles on the rate of progression of vertically transmitted HIV infection in children: association of several HLA-DR13 alleles with long-term survivorship and the potential association of HLA-A*2301 with rapid progression to AIDS. Long-Term Survivor Study. Hum. Immunol. 55:154-162.
- ↵ Critchfield, J. W., D. H. Young, T. L. Hayes, J. V. Braun, J. C. Garcia, R. B. Pollard, and B. L. Shacklett. 2008. Magnitude and complexity of rectal mucosa HIV-1-specific CD8+ T-cell responses during chronic infection reflect clinical status. PLoS One 3:e3577. CrossRefMedline
- ↵ Deeks, S. G., and B. D. Walker. 2007. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27:406-416. CrossRefMedline
- ↵ den Uyl, D., I. E. van der Horst-Bruinsma, and M. van Agtmael. 2004. Progression of HIV to AIDS: a protective role for HLA-B27? AIDS Rev. 6:89-96. Medline
- ↵ Emu, B., E. Sinclair, D. Favre, W. J. Moretto, P. Hsue, R. Hoh, J. N. Martin, D. F. Nixon, J. M. McCune, and S. G. Deeks. 2005. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J. Virol. 79:14169-14178. Abstract/FREE Full Text
- ↵ Emu, B., E. Sinclair, H. Hatano, A. Ferre, B. Shacklett, J. N. Martin, J. M. McCune, and S. G. Deeks. 2008. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J. Virol. 82:5398-5407. Abstract/FREE Full Text
- ↵ Ferre, A. L., P. W. Hunt, J. W. Critchfield, D. H. Young, M. M. Morris, J. C. Garcia, R. B. Pollard, H. F. Yee, Jr., J. N. Martin, S. G. Deeks, and B. L. Shacklett. 2009. Mucosal immune responses to HIV-1 in elite controllers: a potential correlate of immune control. Blood 113:3978-3989. Abstract/FREE Full Text
- ↵ Ferre, A. L., D. Lemongello, P. W. Hunt, M. M. Morris, J. C. Garcia, R. B. Pollard, H. F. Yee, Jr., J. N. Martin, S. G. Deeks, and B. L. Shacklett. 28 July 2010. Immunodominant HIV-specific CD8+ T-cell responses are common to blood and gastrointestinal mucosa, and Gag-specific responses dominate in rectal mucosa of HIV controllers. J. Virol. doi:10.1128/JVI.00803-10. Abstract/FREE Full Text
- ↵ Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. St John, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187-2200. Abstract/FREE Full Text
- ↵ Giraldo-Vela, J. P., R. Rudersdorf, C. Chung, Y. Qi, L. T. Wallace, B. Bimber, G. J. Borchardt, D. L. Fisk, C. E. Glidden, J. T. Loffredo, S. M. Piaskowski, J. R. Furlott, J. P. Morales-Martinez, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2008. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J. Virol. 82:859-870. Abstract/FREE Full Text
- ↵ Goulder, P. J., M. Bunce, P. Krausa, K. McIntyre, S. Crowley, B. Morgan, A. Edwards, P. Giangrande, R. E. Phillips, and A. J. McMichael. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retroviruses 12:1691-1698. Medline
- ↵ Harari, A., S. Petitpierre, F. Vallelian, and G. Pantaleo. 2004. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood 103:966-972. Abstract/FREE Full Text
- ↵ Harari, A., F. Vallelian, P. R. Meylan, and G. Pantaleo. 2005. Functional heterogeneity of memory CD4 T cell responses in different conditions of antigen exposure and persistence. J. Immunol. 174:1037-1045. Abstract/FREE Full Text
- ↵ Hatano, H., E. L. Delwart, P. J. Norris, T. H. Lee, J. Dunn-Williams, P. W. Hunt, R. Hoh, S. L. Stramer, J. M. Linnen, J. M. McCune, J. N. Martin, M. P. Busch, and S. G. Deeks. 2009. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J. Virol. 83:329-335. Abstract/FREE Full Text
- ↵ Hill, A. V., C. E. Allsopp, D. Kwiatkowski, N. M. Anstey, P. Twumasi, P. A. Rowe, S. Bennett, D. Brewster, A. J. McMichael, and B. M. Greenwood. 1991. Common west African HLA antigens are associated with protection from severe malaria. Nature 352:595-600. CrossRefMedline
- ↵ Ibarrondo, F. J., P. A. Anton, M. Fuerst, H. L. Ng, J. T. Wong, J. Matud, J. Elliott, R. Shih, M. A. Hausner, C. Price, L. E. Hultin, P. M. Hultin, B. D. Jamieson, and O. O. Yang. 2005. Parallel human immunodeficiency virus type 1-specific CD8+ T-lymphocyte responses in blood and mucosa during chronic infection. J. Virol. 79:4289-4297. Abstract/FREE Full Text
- ↵ Johnson, R. P., A. Trocha, L. Yang, G. P. Mazzara, D. L. Panicali, T. M. Buchanan, and B. D. Walker. 1991. HIV-1 gag-specific cytotoxic T lymphocytes recognize multiple highly conserved epitopes. Fine specificity of the gag-specific response defined by using unstimulated peripheral blood mononuclear cells and cloned effector cells. J. Immunol. 147:1512-1521. Abstract
- ↵ Juelg, B., E. Moodely, Y. Qi, K. Bishop, S. Reddy, P. J. Goulder, R. Detels, T. Ndung'u, B. D. Walker, and M. Carrington. 2010. HLA class II allele DRB1*1303 is associated with reduced viral loads in 2 independent cohorts of different ethnicity and HIV clades of infection, abstr. 454. In J. W. Mellors, J. M. Coffin, and K. M. De Cock (ed.), 17th Conference on Retroviruses and Opportunistic Infections. CROI, Alexandria, VA. Conference on Retroviruses and Opportunistic Infections, San Francisco, CA.
- ↵ Kalams, S. A., S. P. Buchbinder, E. S. Rosenberg, J. M. Billingsley, D. S. Colbert, N. G. Jones, A. K. Shea, A. K. Trocha, and B. D. Walker. 1999. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J. Virol. 73:6715-6720. Abstract/FREE Full Text
- ↵ Kalams, S. A., and B. D. Walker. 1998. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses [comment]. J. Exp. Med. 188:2199-2204. FREE Full Text
- ↵ Kaufmann, D. E., P. M. Bailey, J. Sidney, B. Wagner, P. J. Norris, M. N. Johnston, L. A. Cosimi, M. M. Addo, M. Lichterfeld, M. Altfeld, N. Frahm, C. Brander, A. Sette, B. D. Walker, and E. S. Rosenberg. 2004. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J. Virol. 78:4463-4477. Abstract/FREE Full Text
- ↵ Keet, I. P. M., J. Tang, M. R. Klein, S. LeBlanc, C. Enger, C. Rivers, R. J. Apple, D. Mann, J. J. Goedert, F. Miedema, and R. A. Kaslow. 1999. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J. Infect. Dis. 180:299-309. Abstract/FREE Full Text
- ↵ Klein, M. R., C. A. van Baalen, A. M. Holwerda, S. R. Kerkhof Garde, R. J. Bende, I. P. Keet, J. K. Eeftinck-Schattenkerk, A. D. Osterhaus, H. Schuitemaker, and F. Miedema. 1995. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp. Med. 181:1365-1372. Abstract/FREE Full Text
- ↵ Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. Abstract/FREE Full Text
- ↵ Lalvani, A., T. Dong, G. Ogg, A. A. Patham, H. Newell, A. V. Hill, A. J. McMichael, and S. Rowland-Jones. 1997. Optimization of a peptide-based protocol employing IL-7 for in vitro restimulation of human cytotoxic T lymphocyte precursors. J. Immunol. Methods 210:65-77. CrossRefMedline
- ↵ Louwagie, J., F. E. McCutchan, M. Peeters, T. P. Brennan, E. Sanders-Buell, G. A. Eddy, G. van der Groen, K. Fransen, G. M. Gershy-Damet, R. Deleys, et al. 1993. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS 7:769-780. Medline
- ↵ Malhotra, U., S. Holte, S. Dutta, M. M. Berrey, E. Delpit, D. M. Koelle, A. Sette, L. Corey, and M. J. McElrath. 2001. Role for HLA class II molecules in HIV-1 suppression and cellular immunity following antiretroviral treatment. J. Clin. Invest. 107:505-517. Medline
- ↵ Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617-3623. Abstract/FREE Full Text
- ↵ Matloubian, M., R. J. Concepcion, and R. Ahmed. 1994. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 68:8056-8063. Abstract/FREE Full Text
- ↵ Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709-2714. Abstract/FREE Full Text
- ↵ Miura, T., M. A. Brockman, A. Schneidewind, M. Lobritz, F. Pereyra, A. Rathod, B. L. Block, Z. L. Brumme, C. J. Brumme, B. Baker, A. C. Rothchild, B. Li, A. Trocha, E. Cutrell, N. Frahm, C. Brander, I. Toth, E. J. Arts, T. M. Allen, and B. D. Walker. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare Gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J. Virol. 83:2743-2755. Abstract/FREE Full Text
- ↵ Nakamura, Y., M. Kameoka, M. Tobiume, M. Kaya, K. Ohki, T. Yamada, and K. Ikuta. 1997. A chain section containing epitopes for cytotoxic T, B and helper T cells within a highly conserved region found in the human immunodeficiency virus type 1 Gag protein. Vaccine 15:489-496. CrossRefMedline
- ↵ Nishino, Y., K. Ohki, T. Kimura, S. Morikawa, T. Mikami, and K. Ikuta. 1992. Major core proteins, p24s, of human, simian, and feline immunodeficiency viruses are partly expressed on the surface of the virus-infected cells. Vaccine 10:677-683. CrossRefMedline
- ↵ Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. Abstract/FREE Full Text
- ↵ Pereyra, F., M. M. Addo, D. E. Kaufmann, Y. Liu, T. Miura, A. Rathod, B. Baker, A. Trocha, R. Rosenberg, E. Mackey, P. Ueda, Z. Lu, D. Cohen, T. Wrin, C. J. Petropoulos, E. S. Rosenberg, and B. D. Walker. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197:563-571. Abstract/FREE Full Text
- ↵ Potter, S. J., C. Lacabaratz, O. Lambotte, S. Perez-Patrigeon, B. Vingert, M. Sinet, J. H. Colle, A. Urrutia, D. Scott-Algara, F. Boufassa, J. F. Delfraissy, J. Theze, A. Venet, and L. A. Chakrabarti. 2007. Preserved central memory and activated effector memory CD4+ T-cell subsets in human immunodeficiency virus controllers: an ANRS EP36 study. J. Virol. 81:13904-13915. Abstract/FREE Full Text
- ↵ Rosenberg, E. S., J. M. Billingsley, A. M. Caliendo, S. L. Boswell, P. E. Sax, S. A. Kalams, and B. D. Walker. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447-1450. Abstract/FREE Full Text
- ↵ Schneidewind, A., M. A. Brockman, J. Sidney, Y. E. Wang, H. Chen, T. J. Suscovich, B. Li, R. I. Adam, R. L. Allgaier, B. R. Mothe, T. Kuntzen, C. Oniangue-Ndza, A. Trocha, X. G. Yu, C. Brander, A. Sette, B. D. Walker, and T. M. Allen. 2008. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 82:5594-5605. Abstract/FREE Full Text
- ↵ Schneidewind, A., M. A. Brockman, R. Yang, R. I. Adam, B. Li, S. Le Gall, C. R. Rinaldo, S. L. Craggs, R. L. Allgaier, K. A. Power, T. Kuntzen, C. S. Tung, M. X. LaBute, S. M. Mueller, T. Harrer, A. J. McMichael, P. J. Goulder, C. Aiken, C. Brander, A. D. Kelleher, and T. M. Allen. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382-12393. Abstract/FREE Full Text
- ↵ Shacklett, B. L., J. W. Critchfield, and D. Lemongello. 2009. Isolating mucosal lymphocytes from biopsy tissue for cellular immunology assays. Methods Mol. Biol. 485:347-356. CrossRefMedline
- ↵ Shacklett, B. L., J. W. Critchfield, and D. Lemongello. 2009. Quantifying HIV-1-specific CD8(+) T-cell responses using ELISPOT and cytokine flow cytometry. Methods Mol. Biol. 485:359-374. CrossRefMedline
- ↵ Shacklett, B. L., O. O. Yang, M. A. Hausner, J. Elliott, L. E. Hultin, C. Price, M. Fuerst, J. Matud, P. Hultin, C. A. Cox, J. Ibarrondo, J. T. Wong, D. F. Nixon, P. A. Anton, and B. D. Jamieson. 2003. Optimization of methods to assess human mucosal T-cell responses to HIV infection and vaccination. J. Immunol. Methods 279:17-31. CrossRefMedline
- ↵ Southwood, S., J. Sidney, A. Kondo, M. F. del Guercio, E. Appella, S. Hoffman, R. T. Kubo, R. W. Chesnut, H. M. Grey, and A. Sette. 1998. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 160:3363-3373. Abstract/FREE Full Text
- ↵ Thursz, M. R., D. Kwiatkowski, C. E. Allsopp, B. M. Greenwood, H. C. Thomas, and A. V. Hill. 1995. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N. Engl. J. Med. 332:1065-1069. CrossRefMedline
- ↵ Vingert, B., S. Perez-Patrigeon, P. Jeannin, O. Lambotte, F. Boufassa, F. Lemaitre, W. W. Kwok, I. Theodorou, J. F. Delfraissy, J. Theze, and L. A. Chakrabarti. 2010. HIV controller CD4+ T cells respond to minimal amounts of Gag antigen due to high TCR avidity. PLoS Pathog. 6:e1000780. CrossRefMedline
- ↵ Vyakarnam, A., D. Sidebottom, S. Murad, J. A. Underhill, P. J. Easterbrook, A. G. Dalgleish, and M. Peakman. 2004. Possession of human leucocyte antigen DQ6 alleles and the rate of CD4 T-cell decline in human immunodeficiency virus-1 infection. Immunology 112:136-142. CrossRefMedline
- ↵ Wilson, J. D., N. Imami, A. Watkins, J. Gill, P. Hay, B. Gazzard, M. Westby, and F. M. Gotch. 2000. Loss of CD4+ T cell proliferative ability but not loss of human immunodeficiency virus type 1 specificity equates with progression to disease. J. Infect. Dis. 182:792-798. Abstract/FREE Full Text
- ↵ Wong, J. T., A. A. Eylath, I. Ghobrial, and R. B. Colvin. 1990. The mechanism of anti-CD3 monoclonal antibodies. Mediation of cytolysis by inter-T cell bridging. Transplantation 50:683-689. CrossRefMedline
- ↵ Zajac, A. J., J. N. Blattman, K. Murali-Krishna, D. J. Sourdive, M. Suresh, J. D. Altman, and R. Ahmed. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205-2213. Abstract/FREE Full Text